Gene
KWMTBOMO04579
Pre Gene Modal
BGIBMGA005480
Annotation
PREDICTED:_vacuolar_fusion_protein_CCZ1_homolog_isoform_X1_[Papilio_xuthus]
Full name
Vacuolar fusion protein CCZ1 homolog
+ More
Vacuolar fusion protein CCZ1 homolog B
Vacuolar fusion protein CCZ1 homolog B
Alternative Name
Vacuolar fusion protein CCZ1 homolog-like
Location in the cell
Cytoplasmic Reliability : 1.169 PlasmaMembrane Reliability : 1.699
Sequence
CDS
ATGATAGACTTAAGATCTAAAATCAACTTTTTCTTCGTTTTTAACCCTACCTTTGGACCTAAGGAGGGCGATGAATTAAAGAGAATACTATACTTTTATCCAAATGAAGTCAGTCCAGATGCCCGGAAAATGCAAGTTGGGCTGTGTGAAGCCGTTGTCAAATTTATGTCGACGTTTTCATCTGAACCATGTGAAGCTTTGCAAACACAATCTAAGAGATACATATTTTTTCAACCAGAAAAAAACTTTTGGATGGTTTTAGTAGTCCGAATACCGTATACAACCAAAGCATTGTCAGCAGTTGGTGAAAGTCAGGGAGATATAGTCAATCCTACTGTAATGCAAAATCTCCTAACATCTGCATATAAGATGTTTAAAATGTTCAGGGGCCCATTCAAAAATATTCCAGCTGATGATTTATATGGTATAAGTGAGCAGTTCTTTACGCCTCTTCTATGGAACGGTCTGTGTACAGAAGACATGCTAACATTATACCAATATCTAGTACAGACATTGCTCCCTAAACAAGTTGAAATAGAAATCCAAGGTGGTGTAGTGACAGCAGCGCAGAGACTTGGCCGATTCATAAGTCCGCCAGATGGAATACATGTTAAAGAAGACTTAGATAAATTGATAAAGATACACGTAACACTCGAAGATGATACAGAATTTAAGGAATACTACCTCCTCATATACAGAACATTGAGTGCGACTGTCTGTTTCACTATTGATGTAAACACTCGGCTGGAACTTGATACGGTCAAGTCAGTGGACGCATTTATTGGGCCGCAGCTATCAGCAATAGCTTCGGCCATCAGCGAACAATGTTCATTGCACGCATTACAAGCAGCGCAACTGGCTAGCGAACACAAGTTCGTGTACTTCAACAGATTAAATTTAATTTTGAAATCATCTCCACCAGTAAATAAATTGCAAATATCCGCTGCGATAAAACCTGAAGTTCTTAGTATCATGTCTGGAATTCATGCTGACCGACAAAGCCTTGGAAATTATGGAGAAATAATAATTAAAACTCCAGACGAATACTGGATAACAGGAAAATGTTCCAATGACAGAGAATTCTATGTGATAATACAAGAGAAAAATGCCAACCTTAAAGAAATAGCGGATGAAGTGAAGCGTGTCTGTGAAGCACAAATGAAAGGGATATTCTTTTACCCTATGTAA
Protein
MIDLRSKINFFFVFNPTFGPKEGDELKRILYFYPNEVSPDARKMQVGLCEAVVKFMSTFSSEPCEALQTQSKRYIFFQPEKNFWMVLVVRIPYTTKALSAVGESQGDIVNPTVMQNLLTSAYKMFKMFRGPFKNIPADDLYGISEQFFTPLLWNGLCTEDMLTLYQYLVQTLLPKQVEIEIQGGVVTAAQRLGRFISPPDGIHVKEDLDKLIKIHVTLEDDTEFKEYYLLIYRTLSATVCFTIDVNTRLELDTVKSVDAFIGPQLSAIASAISEQCSLHALQAAQLASEHKFVYFNRLNLILKSSPPVNKLQISAAIKPEVLSIMSGIHADRQSLGNYGEIIIKTPDEYWITGKCSNDREFYVIIQEKNANLKEIADEVKRVCEAQMKGIFFYPM
Summary
Description
Acts in concert with MON1A, as a guanine exchange factor (GEF) for RAB7, promotes the exchange of GDP to GTP, converting it from an inactive GDP-bound form into an active GTP-bound form.
Acts in concert with MON1A, as a guanine exchange factor (GEF) for RAB7, promotes the exchange of GDP to GTP, converting it from an inactive GDP-bound form into an active GTP-bound form (PubMed:23084991).
Acts in concert with MON1A, as a guanine exchange factor (GEF) for RAB7, promotes the exchange of GDP to GTP, converting it from an inactive GDP-bound form into an active GTP-bound form (PubMed:23084991).
Subunit
Interacts with MON1A. Found in a complex with RMC1, CCZ1 MON1A and MON1B.
Interacts with MON1A. Found in a complex with RMC1, CCZ1, MON1A and MON1B.
Interacts with MON1A (PubMed:23084991). Found in a complex with RMC1, CCZ1, MON1A and MON1B (PubMed:29038162).
Interacts with MON1A. Found in a complex with RMC1, CCZ1, MON1A and MON1B.
Interacts with MON1A (PubMed:23084991). Found in a complex with RMC1, CCZ1, MON1A and MON1B (PubMed:29038162).
Similarity
Belongs to the CCZ1 family.
Keywords
Acetylation
Complete proteome
Guanine-nucleotide releasing factor
Lysosome
Membrane
Phosphoprotein
Reference proteome
Feature
chain Vacuolar fusion protein CCZ1 homolog
Uniprot
A0A194PWR0
A0A194QU31
A0A3S2N9X2
A0A2H1WXG5
H9J7I8
A0A2A4JEW2
+ More
S4NK59 A0A212EL52 A0A0B7A2C9 A0A3L7HQ82 A0A1S3IZ04 A0A067R0N2 D6WEZ5 A0A232FEF7 A0A140UHX9 Q8C1Y8 F7DNC8 A0A1L8EY01 A0A1U7R302 Q28HU2 F7C5J2 F6XPR3 A0A1U7ULN4 A0A1W7RBN4 T1E444 G3IC82 U3FYS6 G1SSD7 E2B812 A0A151N1E6 A0A2K6GIC1 K9ILB0 G3WN95 H0XIL6 A0A1L8EQF7 Q7T102 A0A0B7A1T9 A0A2D4MSQ3 H9GIQ0 A0A250YIR5 G5BF32 G1Q273 I3M786 A0A1B6LR64 Q0VD30 A0A091PT21 W5MPS9 A0A2D4NK71 A0A341BHI4 A0A2U3V336 A0A2Y9P8T5 A0A2Y9SB26 M3X7D0 A0A1V4KS48 A0A087ZSP4 A0A2A3EEL0 F1NPX4 A0A2P4THD5 A0A2Y9J630 Q5ZLN2 A0A226PTA4 A0A093G479 A0A091S9U4 U6D8S8 J3JV42 A0A2Y9DTU4 A0A2J8KJT1 A0A091D499 A0A099ZL34 A0A0Q3XBR0 A0A340WPL3 G1LQY0 A0A3Q7X425 A0A250Y1U2 A0A2K5EJ21 K7BL66 H2QU52 F7AJ87 U3KHS8 A0A3Q7NMA6 H0VRU5 H2RGW5 G2HGQ4 P86791 P86790 A0A2K5PCG3 U3BMT8 A0A2Y9GAB2 H2PLG8 U3BAR2 H9Z9Y4 A0A2K6QQ63 A0A218V7P8 H9H486 H9Z9Y5 H9FVN6 G3TTW3 A0A096NQ31
S4NK59 A0A212EL52 A0A0B7A2C9 A0A3L7HQ82 A0A1S3IZ04 A0A067R0N2 D6WEZ5 A0A232FEF7 A0A140UHX9 Q8C1Y8 F7DNC8 A0A1L8EY01 A0A1U7R302 Q28HU2 F7C5J2 F6XPR3 A0A1U7ULN4 A0A1W7RBN4 T1E444 G3IC82 U3FYS6 G1SSD7 E2B812 A0A151N1E6 A0A2K6GIC1 K9ILB0 G3WN95 H0XIL6 A0A1L8EQF7 Q7T102 A0A0B7A1T9 A0A2D4MSQ3 H9GIQ0 A0A250YIR5 G5BF32 G1Q273 I3M786 A0A1B6LR64 Q0VD30 A0A091PT21 W5MPS9 A0A2D4NK71 A0A341BHI4 A0A2U3V336 A0A2Y9P8T5 A0A2Y9SB26 M3X7D0 A0A1V4KS48 A0A087ZSP4 A0A2A3EEL0 F1NPX4 A0A2P4THD5 A0A2Y9J630 Q5ZLN2 A0A226PTA4 A0A093G479 A0A091S9U4 U6D8S8 J3JV42 A0A2Y9DTU4 A0A2J8KJT1 A0A091D499 A0A099ZL34 A0A0Q3XBR0 A0A340WPL3 G1LQY0 A0A3Q7X425 A0A250Y1U2 A0A2K5EJ21 K7BL66 H2QU52 F7AJ87 U3KHS8 A0A3Q7NMA6 H0VRU5 H2RGW5 G2HGQ4 P86791 P86790 A0A2K5PCG3 U3BMT8 A0A2Y9GAB2 H2PLG8 U3BAR2 H9Z9Y4 A0A2K6QQ63 A0A218V7P8 H9H486 H9Z9Y5 H9FVN6 G3TTW3 A0A096NQ31
Pubmed
26354079
19121390
23622113
22118469
29704459
24845553
+ More
18362917 19820115 28648823 15057822 16141072 15489334 19144319 21183079 20431018 27762356 18464734 26358130 23758969 21804562 23915248 21993624 20798317 22293439 21709235 21881562 28087693 21993625 17975172 15592404 15642098 24621616 22516182 20010809 16136131 19892987 21484476 10810093 14702039 12853948 17897319 18220336 18669648 19413330 21269460 23084991 22223895 23186163 29038162 25243066 25319552 25362486 17431167
18362917 19820115 28648823 15057822 16141072 15489334 19144319 21183079 20431018 27762356 18464734 26358130 23758969 21804562 23915248 21993624 20798317 22293439 21709235 21881562 28087693 21993625 17975172 15592404 15642098 24621616 22516182 20010809 16136131 19892987 21484476 10810093 14702039 12853948 17897319 18220336 18669648 19413330 21269460 23084991 22223895 23186163 29038162 25243066 25319552 25362486 17431167
EMBL
KQ459595
KPI95570.1
KQ461154
KPJ08500.1
RSAL01000163
RVE45368.1
+ More
ODYU01011808 SOQ57765.1 BABH01013921 NWSH01001841 PCG69942.1 GAIX01013429 JAA79131.1 AGBW02014132 OWR42213.1 HACG01027917 CEK74782.1 RAZU01000163 RLQ68066.1 KK852824 KDR15453.1 KQ971319 EFA00460.2 NNAY01000365 OXU28858.1 AC126486 AK090008 AK150793 AK152328 AK152576 AK153160 BC048077 AAMC01007054 AAMC01007055 AAMC01007056 CM004482 OCT64210.1 CR760737 GDAY02002918 JAV48549.1 GAAZ01002916 JAA95027.1 JH001891 EGW02542.1 GAEP01001871 JAB52950.1 GL446270 EFN88171.1 AKHW03004154 KYO30646.1 GABZ01005828 JAA47697.1 AEFK01075610 AEFK01075611 AEFK01075612 AEFK01075613 AEFK01075614 AEFK01075615 AAQR03148892 AAQR03148893 AAQR03148894 AAQR03148895 CM004483 OCT61586.1 BC055965 HACG01027918 CEK74783.1 IACM01117687 LAB35803.1 AAWZ02029387 GFFW01001260 JAV43528.1 JH169911 GEBF01002442 EHB07893.1 JAO01191.1 AAPE02034763 AGTP01102874 GEBQ01013802 JAT26175.1 BC119863 KK678304 KFQ10810.1 AHAT01013702 IACN01006609 LAB46094.1 AANG04004562 LSYS01001700 OPJ87253.1 KZ288266 PBC30148.1 AADN05000376 PPHD01000222 POI35765.1 AJ719702 AWGT02000022 OXB82712.1 KL214745 KFV61602.1 KK813486 KFQ37220.1 HAAF01006484 CCP78308.1 BT127108 AEE62070.1 NBAG03000362 PNI35261.1 KN123370 KFO25323.1 KL896048 KGL83154.1 LMAW01000001 KQL61533.1 ACTA01178351 ACTA01186349 GFFV01002420 JAV37525.1 GABC01000765 GABF01007286 JAA10573.1 JAA14859.1 AC193266 GABC01000764 GABF01007285 GABD01003299 GABD01003298 GABE01009998 JAA10574.1 JAA14860.1 JAA29801.1 JAA34741.1 AGTO01020915 AAKN02032157 AAKN02032158 AAKN02032159 AC192442 AK305918 BAK62912.1 AF151801 AK292326 AC004983 BC132749 BC132751 AC079882 BC010130 GAMS01008908 GAMR01004692 GAMQ01005809 GAMP01002176 JAB14228.1 JAB29240.1 JAB36042.1 JAB50579.1 ABGA01163992 ABGA01163993 NDHI03004471 PNJ01525.1 GAMT01008599 JAB03262.1 JU475946 AFH32750.1 MUZQ01000034 OWK61949.1 JSUE03027732 JSUE03027733 JSUE03027734 JSUE03027735 JSUE03027736 JSUE03027737 JSUE03027738 JSUE03027739 JSUE03027740 JSUE03027741 JU475947 AFH32751.1 JU334942 AFE78695.1 AHZZ02022003
ODYU01011808 SOQ57765.1 BABH01013921 NWSH01001841 PCG69942.1 GAIX01013429 JAA79131.1 AGBW02014132 OWR42213.1 HACG01027917 CEK74782.1 RAZU01000163 RLQ68066.1 KK852824 KDR15453.1 KQ971319 EFA00460.2 NNAY01000365 OXU28858.1 AC126486 AK090008 AK150793 AK152328 AK152576 AK153160 BC048077 AAMC01007054 AAMC01007055 AAMC01007056 CM004482 OCT64210.1 CR760737 GDAY02002918 JAV48549.1 GAAZ01002916 JAA95027.1 JH001891 EGW02542.1 GAEP01001871 JAB52950.1 GL446270 EFN88171.1 AKHW03004154 KYO30646.1 GABZ01005828 JAA47697.1 AEFK01075610 AEFK01075611 AEFK01075612 AEFK01075613 AEFK01075614 AEFK01075615 AAQR03148892 AAQR03148893 AAQR03148894 AAQR03148895 CM004483 OCT61586.1 BC055965 HACG01027918 CEK74783.1 IACM01117687 LAB35803.1 AAWZ02029387 GFFW01001260 JAV43528.1 JH169911 GEBF01002442 EHB07893.1 JAO01191.1 AAPE02034763 AGTP01102874 GEBQ01013802 JAT26175.1 BC119863 KK678304 KFQ10810.1 AHAT01013702 IACN01006609 LAB46094.1 AANG04004562 LSYS01001700 OPJ87253.1 KZ288266 PBC30148.1 AADN05000376 PPHD01000222 POI35765.1 AJ719702 AWGT02000022 OXB82712.1 KL214745 KFV61602.1 KK813486 KFQ37220.1 HAAF01006484 CCP78308.1 BT127108 AEE62070.1 NBAG03000362 PNI35261.1 KN123370 KFO25323.1 KL896048 KGL83154.1 LMAW01000001 KQL61533.1 ACTA01178351 ACTA01186349 GFFV01002420 JAV37525.1 GABC01000765 GABF01007286 JAA10573.1 JAA14859.1 AC193266 GABC01000764 GABF01007285 GABD01003299 GABD01003298 GABE01009998 JAA10574.1 JAA14860.1 JAA29801.1 JAA34741.1 AGTO01020915 AAKN02032157 AAKN02032158 AAKN02032159 AC192442 AK305918 BAK62912.1 AF151801 AK292326 AC004983 BC132749 BC132751 AC079882 BC010130 GAMS01008908 GAMR01004692 GAMQ01005809 GAMP01002176 JAB14228.1 JAB29240.1 JAB36042.1 JAB50579.1 ABGA01163992 ABGA01163993 NDHI03004471 PNJ01525.1 GAMT01008599 JAB03262.1 JU475946 AFH32750.1 MUZQ01000034 OWK61949.1 JSUE03027732 JSUE03027733 JSUE03027734 JSUE03027735 JSUE03027736 JSUE03027737 JSUE03027738 JSUE03027739 JSUE03027740 JSUE03027741 JU475947 AFH32751.1 JU334942 AFE78695.1 AHZZ02022003
Proteomes
UP000053268
UP000053240
UP000283053
UP000005204
UP000218220
UP000007151
+ More
UP000273346 UP000085678 UP000027135 UP000007266 UP000215335 UP000002494 UP000000589 UP000008143 UP000186698 UP000189706 UP000002279 UP000189704 UP000001075 UP000001811 UP000008237 UP000050525 UP000233160 UP000007648 UP000005225 UP000001646 UP000006813 UP000001074 UP000005215 UP000009136 UP000018468 UP000252040 UP000245320 UP000248483 UP000248484 UP000011712 UP000190648 UP000005203 UP000242457 UP000000539 UP000248482 UP000198419 UP000053875 UP000248480 UP000028990 UP000053641 UP000051836 UP000265300 UP000008912 UP000286642 UP000233020 UP000002277 UP000002281 UP000016665 UP000286641 UP000005447 UP000005640 UP000233040 UP000008225 UP000248481 UP000001595 UP000233200 UP000197619 UP000006718 UP000007646 UP000028761
UP000273346 UP000085678 UP000027135 UP000007266 UP000215335 UP000002494 UP000000589 UP000008143 UP000186698 UP000189706 UP000002279 UP000189704 UP000001075 UP000001811 UP000008237 UP000050525 UP000233160 UP000007648 UP000005225 UP000001646 UP000006813 UP000001074 UP000005215 UP000009136 UP000018468 UP000252040 UP000245320 UP000248483 UP000248484 UP000011712 UP000190648 UP000005203 UP000242457 UP000000539 UP000248482 UP000198419 UP000053875 UP000248480 UP000028990 UP000053641 UP000051836 UP000265300 UP000008912 UP000286642 UP000233020 UP000002277 UP000002281 UP000016665 UP000286641 UP000005447 UP000005640 UP000233040 UP000008225 UP000248481 UP000001595 UP000233200 UP000197619 UP000006718 UP000007646 UP000028761
Pfam
PF08217 DUF1712
Interpro
IPR013176
Ccz1
ProteinModelPortal
A0A194PWR0
A0A194QU31
A0A3S2N9X2
A0A2H1WXG5
H9J7I8
A0A2A4JEW2
+ More
S4NK59 A0A212EL52 A0A0B7A2C9 A0A3L7HQ82 A0A1S3IZ04 A0A067R0N2 D6WEZ5 A0A232FEF7 A0A140UHX9 Q8C1Y8 F7DNC8 A0A1L8EY01 A0A1U7R302 Q28HU2 F7C5J2 F6XPR3 A0A1U7ULN4 A0A1W7RBN4 T1E444 G3IC82 U3FYS6 G1SSD7 E2B812 A0A151N1E6 A0A2K6GIC1 K9ILB0 G3WN95 H0XIL6 A0A1L8EQF7 Q7T102 A0A0B7A1T9 A0A2D4MSQ3 H9GIQ0 A0A250YIR5 G5BF32 G1Q273 I3M786 A0A1B6LR64 Q0VD30 A0A091PT21 W5MPS9 A0A2D4NK71 A0A341BHI4 A0A2U3V336 A0A2Y9P8T5 A0A2Y9SB26 M3X7D0 A0A1V4KS48 A0A087ZSP4 A0A2A3EEL0 F1NPX4 A0A2P4THD5 A0A2Y9J630 Q5ZLN2 A0A226PTA4 A0A093G479 A0A091S9U4 U6D8S8 J3JV42 A0A2Y9DTU4 A0A2J8KJT1 A0A091D499 A0A099ZL34 A0A0Q3XBR0 A0A340WPL3 G1LQY0 A0A3Q7X425 A0A250Y1U2 A0A2K5EJ21 K7BL66 H2QU52 F7AJ87 U3KHS8 A0A3Q7NMA6 H0VRU5 H2RGW5 G2HGQ4 P86791 P86790 A0A2K5PCG3 U3BMT8 A0A2Y9GAB2 H2PLG8 U3BAR2 H9Z9Y4 A0A2K6QQ63 A0A218V7P8 H9H486 H9Z9Y5 H9FVN6 G3TTW3 A0A096NQ31
S4NK59 A0A212EL52 A0A0B7A2C9 A0A3L7HQ82 A0A1S3IZ04 A0A067R0N2 D6WEZ5 A0A232FEF7 A0A140UHX9 Q8C1Y8 F7DNC8 A0A1L8EY01 A0A1U7R302 Q28HU2 F7C5J2 F6XPR3 A0A1U7ULN4 A0A1W7RBN4 T1E444 G3IC82 U3FYS6 G1SSD7 E2B812 A0A151N1E6 A0A2K6GIC1 K9ILB0 G3WN95 H0XIL6 A0A1L8EQF7 Q7T102 A0A0B7A1T9 A0A2D4MSQ3 H9GIQ0 A0A250YIR5 G5BF32 G1Q273 I3M786 A0A1B6LR64 Q0VD30 A0A091PT21 W5MPS9 A0A2D4NK71 A0A341BHI4 A0A2U3V336 A0A2Y9P8T5 A0A2Y9SB26 M3X7D0 A0A1V4KS48 A0A087ZSP4 A0A2A3EEL0 F1NPX4 A0A2P4THD5 A0A2Y9J630 Q5ZLN2 A0A226PTA4 A0A093G479 A0A091S9U4 U6D8S8 J3JV42 A0A2Y9DTU4 A0A2J8KJT1 A0A091D499 A0A099ZL34 A0A0Q3XBR0 A0A340WPL3 G1LQY0 A0A3Q7X425 A0A250Y1U2 A0A2K5EJ21 K7BL66 H2QU52 F7AJ87 U3KHS8 A0A3Q7NMA6 H0VRU5 H2RGW5 G2HGQ4 P86791 P86790 A0A2K5PCG3 U3BMT8 A0A2Y9GAB2 H2PLG8 U3BAR2 H9Z9Y4 A0A2K6QQ63 A0A218V7P8 H9H486 H9Z9Y5 H9FVN6 G3TTW3 A0A096NQ31
Ontologies
PANTHER
Topology
Subcellular location
Lysosome membrane
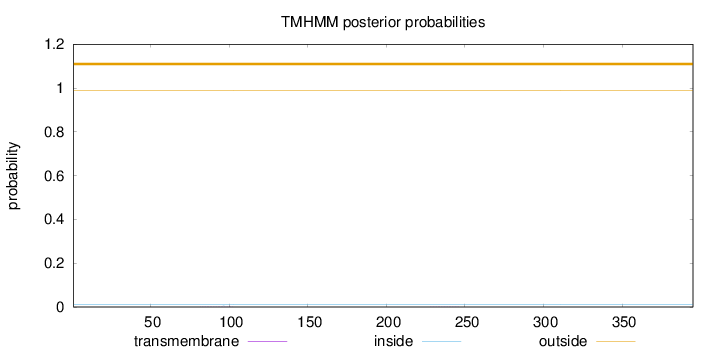
Length:
395
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01385
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.01027
outside
1 - 395
Population Genetic Test Statistics
Pi
228.721983
Theta
150.970328
Tajima's D
1.440116
CLR
0
CSRT
0.773061346932653
Interpretation
Uncertain
