Gene
KWMTBOMO04578
Pre Gene Modal
BGIBMGA005330
Annotation
hypothetical_protein_KGM_22219_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 2.777
Sequence
CDS
ATGGCTATTAGAAATCCCCGCCTTCCGCTGTTACCTGGCTATGGCACTAACCCGCTGCTAGGAAAAAAGAATTTCGGAGTACGTCCAGTATTTGCATCTATTGACAAAGTCAATATGCTAGTTGACAAAGCTGAAGGCGTGAATAAAGTTCCGTCGATGTACAGTCGGAAACAGGCACCTGATCTTCCAACTTGGATCATGTATGATAAGAACATTTTACGGTTTCAAGGGTTCTTTCAACAATCACTGCATGAAATGCGGTCAGCATCTCATATTTTACGTAAAGTTGATATTTTCTTTTTTCTCGAAGATGGTACTATAAAGGTTATAGAGCCGAAGACTGAGAATAGCGGATTGTCGCAAGGTACTTTGATAAGTCGCCAAAGAATACGTTTGCCTTTCAGTTACGACATGTACTATGATGTGCTTGATTTAAACATTGGCCGTGAAGTTACATTTTATGGAAAAACTTTTAAGATTGTAAACTGTGATAATTTTACAAGAGTATTTTTGAATCGTCTTGGTATAAACGTACCTGATCCTATTCCGTGGCCCGATGCAATTGAGAGATCACCTGCCGGTGGAAAACCTCCGAAACAGCGACCTTTTAAGCAATTCCTGGATTATGACAGACAAGTTTTAAGGTTTTACGGATACTGGGATGATCGGGATTCAGAATTTGGTATATTACACAACATGGAAATTCATTACTTTCTGGCGGATGATACCATCGAAATAAAGGAAGTCATGCCAGCCAATTCTGGAATGGAAGCCGGCCCTATGTTCTTGAAGAGAATGCGGTTACCACGAAAAATACCTTCGCATGTCGAAATGACGGGTGGTCCGAAGCAGGCCTCTTATGGTCCGGCGGATCTAAGCATCGGTGCTATTTTAAACGTTTTTGGTCGTAATGTAGTTTTGACCGACTGTGATCCATTCACTAAAGAATATTACAGAGTTACATACGGATTTGACACTTTCACACCGTTGACAATACCAAACAAGGATAGTTCAGAATGTGTGAGCACTAATGTATCAGAACGGAAGTTGCCTCCATGGAACGGCTACGGATCCTACGACGATTCAGCTGAGAACTGTCGTACTGTGGAACCGAAGGCACCTCATAGGGACTTTATGAAATTTATTCATAAAGACAGGGTCGGCTTTGATTCTCATATTCTTCGATTTGCTGCTCGACTCATAACCGACAACCCTGAAGATGAACGCCGCTATTTTATTGTTAAATACTTTTTGTGTGATGATACAATTGGTATCTTTGAGTTGGGCGAAAGGAACTCCGGTTTCAAGGGTGGTAAATTCTTTCGACGCGACAAGATGTATCTGCCAGACGTTGAATTTTTCGTTCCGAACGAACCGCCCGCGTACACAGACAAAGACATGTGGGTCGGAAACGAGCTGGTCATCAACAAGCATCGTTTTCGTCTCATTTCCGCTGACGAATACGCTTTGCGATATTTAGAATTGCATTCGGACGAGTATCCTATGGCGAACATAGCTTTGATTATGGACAAGATTCGAAGGTTTTTGGCGTCTCAAGAAAATGGTTATAAAAACTTTGTGGCTAAATACATGGAAGCTGTTTTACCAAATAATAAGGAACTTATGAGCGTCCGATGTTTCAAGCAAGCTCTGAAGGAAATAATGTGCGAAAAAATGACACAACACGAGTTTTTGACTTTGGTTAGACATTTTCGAGGTGATCCAGGGAAAGAGAAAAGCCCGCGACGTGAAATGATTAGGTCGTTAGTGTTTACGGAGTTGACCCGTGGCCTGTGGGACGACCGCGCCCGGCTGCGGGAGGGGCTCGTGCACGCCGACGAGCTCGGCACCGGGGCGGTGGCGCCGGAGCGCCTGCGACAGCTGCTCCGCGCCCACCGACTCACTATCAACACCGACCTCATGGACTGCATGCTGCAAGTATTGGAAAAGGACGAAAACTGTAACATTTACTATGAAGATTTGTTGAAGTTTTTGGATTTCCAAACTCGACCAGTGTTCAATCTGACGGAGGATGATTATCAGAAGGTCGTCAGGCACGCGCCACCGCTGAAAGATACTGAGTGCAAATTGTGGGCCGAAAACGAGAATCTCATCCAGGAAGGCTACGTCAACTGGAATGCATTCCTGTGCCAACTCAACTTGGAACATCTTGTGAAAGAACAATCTTAG
Protein
MAIRNPRLPLLPGYGTNPLLGKKNFGVRPVFASIDKVNMLVDKAEGVNKVPSMYSRKQAPDLPTWIMYDKNILRFQGFFQQSLHEMRSASHILRKVDIFFFLEDGTIKVIEPKTENSGLSQGTLISRQRIRLPFSYDMYYDVLDLNIGREVTFYGKTFKIVNCDNFTRVFLNRLGINVPDPIPWPDAIERSPAGGKPPKQRPFKQFLDYDRQVLRFYGYWDDRDSEFGILHNMEIHYFLADDTIEIKEVMPANSGMEAGPMFLKRMRLPRKIPSHVEMTGGPKQASYGPADLSIGAILNVFGRNVVLTDCDPFTKEYYRVTYGFDTFTPLTIPNKDSSECVSTNVSERKLPPWNGYGSYDDSAENCRTVEPKAPHRDFMKFIHKDRVGFDSHILRFAARLITDNPEDERRYFIVKYFLCDDTIGIFELGERNSGFKGGKFFRRDKMYLPDVEFFVPNEPPAYTDKDMWVGNELVINKHRFRLISADEYALRYLELHSDEYPMANIALIMDKIRRFLASQENGYKNFVAKYMEAVLPNNKELMSVRCFKQALKEIMCEKMTQHEFLTLVRHFRGDPGKEKSPRREMIRSLVFTELTRGLWDDRARLREGLVHADELGTGAVAPERLRQLLRAHRLTINTDLMDCMLQVLEKDENCNIYYEDLLKFLDFQTRPVFNLTEDDYQKVVRHAPPLKDTECKLWAENENLIQEGYVNWNAFLCQLNLEHLVKEQS
Summary
Uniprot
A0A3S2P9R1
A0A212EL50
A0A194QT54
A0A194PRJ0
H9J738
A0A194PS39
+ More
A0A0L7LD72 D6WLV6 A0A1W4WPK9 A0A3L8E1X8 A0A026W5A2 A0A088A094 A0A154PE17 A0A195CE45 A0A2A3E3U9 A0A0L7QTU2 A0A0M9A7P9 A0A195BW29 A0A151J6C1 A0A151WFM4 A0A158NC99 A0A195F9J5 A0A1Y1KUM1 A0A1I8N6P8 A0A1Q3G5D4 A0A0C9R3K8 W5JNY1 A0A067R4J3 A0A1A9W0X8 A0A336K9S5 A0A336MSJ8 A0A182QI23 A0A084VC23 B3N0M8 A0A182FP82 A0A182J999 A0A182P8P4 A0A182VXS3 Q5TSN7 A0A182HJK3 A0A182TLD2 A0A182XKE1 A0A182KWQ3 A0A182VKP4 A0A1Y1KS54 A0A182RQQ4 A0A1S4FN40 Q16V31 K7IVS8 A0A182XXB1 A0A182K9T0 B4PX67 A0A1W3JUC2 A0A232F7G8 A0A182MCH4 H2ZJ45 A0A1A9ZYI2 A0A1S3JUL5 A0A1S3KBE9 V4BGM1 A0A182NFU6 A0A182SXL3 A0A2P8XPV8 W4Y2G3 A0A1B0G4P1 A0A0M4EV90 Q32TF6 C3ZDQ1 Q28X04 A0A3Q3LX26 B4MAX2 B4H4U8 B4JYU2 B4L5F9 W5M1L2 A0A3Q3LX21 A0A0R3U2Y8 A0A3P9Q8S6 A0A3B4UJD2 A0A3B4UIV3 A0A210QEN1 A0A3B4EF34 A0A3B5B1C8 A0A2B4SEE0 A0A2G8LEF1 B3MGS7 H3AR71 A7SUE4 A0A0X3PC41 A0A1Y1KSM3 G4V5T5 A0A095APE5 W5K4R5 A0A3Q0KSQ7 A0A3P9AL42 B5DEU8 A0A3P9AL37
A0A0L7LD72 D6WLV6 A0A1W4WPK9 A0A3L8E1X8 A0A026W5A2 A0A088A094 A0A154PE17 A0A195CE45 A0A2A3E3U9 A0A0L7QTU2 A0A0M9A7P9 A0A195BW29 A0A151J6C1 A0A151WFM4 A0A158NC99 A0A195F9J5 A0A1Y1KUM1 A0A1I8N6P8 A0A1Q3G5D4 A0A0C9R3K8 W5JNY1 A0A067R4J3 A0A1A9W0X8 A0A336K9S5 A0A336MSJ8 A0A182QI23 A0A084VC23 B3N0M8 A0A182FP82 A0A182J999 A0A182P8P4 A0A182VXS3 Q5TSN7 A0A182HJK3 A0A182TLD2 A0A182XKE1 A0A182KWQ3 A0A182VKP4 A0A1Y1KS54 A0A182RQQ4 A0A1S4FN40 Q16V31 K7IVS8 A0A182XXB1 A0A182K9T0 B4PX67 A0A1W3JUC2 A0A232F7G8 A0A182MCH4 H2ZJ45 A0A1A9ZYI2 A0A1S3JUL5 A0A1S3KBE9 V4BGM1 A0A182NFU6 A0A182SXL3 A0A2P8XPV8 W4Y2G3 A0A1B0G4P1 A0A0M4EV90 Q32TF6 C3ZDQ1 Q28X04 A0A3Q3LX26 B4MAX2 B4H4U8 B4JYU2 B4L5F9 W5M1L2 A0A3Q3LX21 A0A0R3U2Y8 A0A3P9Q8S6 A0A3B4UJD2 A0A3B4UIV3 A0A210QEN1 A0A3B4EF34 A0A3B5B1C8 A0A2B4SEE0 A0A2G8LEF1 B3MGS7 H3AR71 A7SUE4 A0A0X3PC41 A0A1Y1KSM3 G4V5T5 A0A095APE5 W5K4R5 A0A3Q0KSQ7 A0A3P9AL42 B5DEU8 A0A3P9AL37
Pubmed
22118469
26354079
19121390
26227816
18362917
19820115
+ More
30249741 24508170 21347285 28004739 25315136 20920257 23761445 24845553 24438588 17994087 12364791 14747013 17210077 17510324 20075255 25244985 17550304 28648823 23254933 29403074 12481130 15114417 18563158 15632085 28812685 29023486 9215903 17615350 22253936 22246508 25329095 25069045
30249741 24508170 21347285 28004739 25315136 20920257 23761445 24845553 24438588 17994087 12364791 14747013 17210077 17510324 20075255 25244985 17550304 28648823 23254933 29403074 12481130 15114417 18563158 15632085 28812685 29023486 9215903 17615350 22253936 22246508 25329095 25069045
EMBL
RSAL01000163
RVE45367.1
AGBW02014132
OWR42212.1
KQ461154
KPJ08499.1
+ More
KQ459595 KPI95568.1 BABH01013920 BABH01013921 KPI95569.1 JTDY01001597 KOB73422.1 KQ971343 EFA03395.1 QOIP01000001 RLU26667.1 KK107468 EZA50209.1 KQ434885 KZC10057.1 KQ977873 KYM99144.1 KZ288387 PBC26433.1 KQ414740 KOC62040.1 KQ435719 KOX78940.1 KQ976396 KYM92839.1 KQ979863 KYN18767.1 KQ983211 KYQ46627.1 ADTU01011774 KQ981727 KYN37113.1 GEZM01077044 JAV63415.1 GFDL01000038 JAV35007.1 GBYB01007377 JAG77144.1 ADMH02000799 ETN65018.1 KK852751 KDR17132.1 UFQS01000152 UFQT01000152 SSX00552.1 SSX20932.1 UFQS01002315 UFQT01002315 SSX13785.1 SSX33206.1 AXCN02000248 ATLV01009983 KE524560 KFB35517.1 CH902642 EDV34107.1 AAAB01008904 EAL40432.4 APCN01002178 GEZM01077042 GEZM01077028 JAV63418.1 CH477605 EAT38397.1 CM000162 EDX01830.1 NNAY01000843 OXU26237.1 AXCM01001599 KB199651 ESP05007.1 PYGN01001564 PSN34041.1 AAGJ04068632 CCAG010021607 CP012528 ALC49838.1 AY823395 AAX08050.1 GG666612 EEN48857.1 CM000071 EAL26512.2 CH940655 EDW66381.1 CH479210 EDW32784.1 CH916378 EDV98557.1 CH933811 EDW06418.1 AHAT01025095 AHAT01025096 UXSR01000091 VDD74878.1 NEDP02004019 OWF47194.1 LSMT01000117 PFX26805.1 MRZV01000110 PIK58530.1 CH902619 EDV36835.1 AFYH01119873 AFYH01119874 AFYH01119875 AFYH01119876 AFYH01119877 DS469812 EDO32661.1 GEEE01013734 JAP49491.1 GEZM01077030 GEZM01077022 JAV63441.1 HE601624 CCD74648.1 KL250749 KGB36156.1 BC168809 AAI68809.1
KQ459595 KPI95568.1 BABH01013920 BABH01013921 KPI95569.1 JTDY01001597 KOB73422.1 KQ971343 EFA03395.1 QOIP01000001 RLU26667.1 KK107468 EZA50209.1 KQ434885 KZC10057.1 KQ977873 KYM99144.1 KZ288387 PBC26433.1 KQ414740 KOC62040.1 KQ435719 KOX78940.1 KQ976396 KYM92839.1 KQ979863 KYN18767.1 KQ983211 KYQ46627.1 ADTU01011774 KQ981727 KYN37113.1 GEZM01077044 JAV63415.1 GFDL01000038 JAV35007.1 GBYB01007377 JAG77144.1 ADMH02000799 ETN65018.1 KK852751 KDR17132.1 UFQS01000152 UFQT01000152 SSX00552.1 SSX20932.1 UFQS01002315 UFQT01002315 SSX13785.1 SSX33206.1 AXCN02000248 ATLV01009983 KE524560 KFB35517.1 CH902642 EDV34107.1 AAAB01008904 EAL40432.4 APCN01002178 GEZM01077042 GEZM01077028 JAV63418.1 CH477605 EAT38397.1 CM000162 EDX01830.1 NNAY01000843 OXU26237.1 AXCM01001599 KB199651 ESP05007.1 PYGN01001564 PSN34041.1 AAGJ04068632 CCAG010021607 CP012528 ALC49838.1 AY823395 AAX08050.1 GG666612 EEN48857.1 CM000071 EAL26512.2 CH940655 EDW66381.1 CH479210 EDW32784.1 CH916378 EDV98557.1 CH933811 EDW06418.1 AHAT01025095 AHAT01025096 UXSR01000091 VDD74878.1 NEDP02004019 OWF47194.1 LSMT01000117 PFX26805.1 MRZV01000110 PIK58530.1 CH902619 EDV36835.1 AFYH01119873 AFYH01119874 AFYH01119875 AFYH01119876 AFYH01119877 DS469812 EDO32661.1 GEEE01013734 JAP49491.1 GEZM01077030 GEZM01077022 JAV63441.1 HE601624 CCD74648.1 KL250749 KGB36156.1 BC168809 AAI68809.1
Proteomes
UP000283053
UP000007151
UP000053240
UP000053268
UP000005204
UP000037510
+ More
UP000007266 UP000192223 UP000279307 UP000053097 UP000005203 UP000076502 UP000078542 UP000242457 UP000053825 UP000053105 UP000078540 UP000078492 UP000075809 UP000005205 UP000078541 UP000095301 UP000000673 UP000027135 UP000091820 UP000075886 UP000030765 UP000007801 UP000069272 UP000075880 UP000075885 UP000075920 UP000007062 UP000075840 UP000075902 UP000076407 UP000075903 UP000008820 UP000002358 UP000076408 UP000075881 UP000002282 UP000215335 UP000075883 UP000007875 UP000092445 UP000085678 UP000030746 UP000075884 UP000075901 UP000245037 UP000007110 UP000092444 UP000092553 UP000008144 UP000001554 UP000001819 UP000261640 UP000008792 UP000008744 UP000001070 UP000009192 UP000018468 UP000046399 UP000267029 UP000242638 UP000261420 UP000242188 UP000261440 UP000261400 UP000225706 UP000230750 UP000008672 UP000001593 UP000008854 UP000018467 UP000265140
UP000007266 UP000192223 UP000279307 UP000053097 UP000005203 UP000076502 UP000078542 UP000242457 UP000053825 UP000053105 UP000078540 UP000078492 UP000075809 UP000005205 UP000078541 UP000095301 UP000000673 UP000027135 UP000091820 UP000075886 UP000030765 UP000007801 UP000069272 UP000075880 UP000075885 UP000075920 UP000007062 UP000075840 UP000075902 UP000076407 UP000075903 UP000008820 UP000002358 UP000076408 UP000075881 UP000002282 UP000215335 UP000075883 UP000007875 UP000092445 UP000085678 UP000030746 UP000075884 UP000075901 UP000245037 UP000007110 UP000092444 UP000092553 UP000008144 UP000001554 UP000001819 UP000261640 UP000008792 UP000008744 UP000001070 UP000009192 UP000018468 UP000046399 UP000267029 UP000242638 UP000261420 UP000242188 UP000261440 UP000261400 UP000225706 UP000230750 UP000008672 UP000001593 UP000008854 UP000018467 UP000265140
PRIDE
Interpro
ProteinModelPortal
A0A3S2P9R1
A0A212EL50
A0A194QT54
A0A194PRJ0
H9J738
A0A194PS39
+ More
A0A0L7LD72 D6WLV6 A0A1W4WPK9 A0A3L8E1X8 A0A026W5A2 A0A088A094 A0A154PE17 A0A195CE45 A0A2A3E3U9 A0A0L7QTU2 A0A0M9A7P9 A0A195BW29 A0A151J6C1 A0A151WFM4 A0A158NC99 A0A195F9J5 A0A1Y1KUM1 A0A1I8N6P8 A0A1Q3G5D4 A0A0C9R3K8 W5JNY1 A0A067R4J3 A0A1A9W0X8 A0A336K9S5 A0A336MSJ8 A0A182QI23 A0A084VC23 B3N0M8 A0A182FP82 A0A182J999 A0A182P8P4 A0A182VXS3 Q5TSN7 A0A182HJK3 A0A182TLD2 A0A182XKE1 A0A182KWQ3 A0A182VKP4 A0A1Y1KS54 A0A182RQQ4 A0A1S4FN40 Q16V31 K7IVS8 A0A182XXB1 A0A182K9T0 B4PX67 A0A1W3JUC2 A0A232F7G8 A0A182MCH4 H2ZJ45 A0A1A9ZYI2 A0A1S3JUL5 A0A1S3KBE9 V4BGM1 A0A182NFU6 A0A182SXL3 A0A2P8XPV8 W4Y2G3 A0A1B0G4P1 A0A0M4EV90 Q32TF6 C3ZDQ1 Q28X04 A0A3Q3LX26 B4MAX2 B4H4U8 B4JYU2 B4L5F9 W5M1L2 A0A3Q3LX21 A0A0R3U2Y8 A0A3P9Q8S6 A0A3B4UJD2 A0A3B4UIV3 A0A210QEN1 A0A3B4EF34 A0A3B5B1C8 A0A2B4SEE0 A0A2G8LEF1 B3MGS7 H3AR71 A7SUE4 A0A0X3PC41 A0A1Y1KSM3 G4V5T5 A0A095APE5 W5K4R5 A0A3Q0KSQ7 A0A3P9AL42 B5DEU8 A0A3P9AL37
A0A0L7LD72 D6WLV6 A0A1W4WPK9 A0A3L8E1X8 A0A026W5A2 A0A088A094 A0A154PE17 A0A195CE45 A0A2A3E3U9 A0A0L7QTU2 A0A0M9A7P9 A0A195BW29 A0A151J6C1 A0A151WFM4 A0A158NC99 A0A195F9J5 A0A1Y1KUM1 A0A1I8N6P8 A0A1Q3G5D4 A0A0C9R3K8 W5JNY1 A0A067R4J3 A0A1A9W0X8 A0A336K9S5 A0A336MSJ8 A0A182QI23 A0A084VC23 B3N0M8 A0A182FP82 A0A182J999 A0A182P8P4 A0A182VXS3 Q5TSN7 A0A182HJK3 A0A182TLD2 A0A182XKE1 A0A182KWQ3 A0A182VKP4 A0A1Y1KS54 A0A182RQQ4 A0A1S4FN40 Q16V31 K7IVS8 A0A182XXB1 A0A182K9T0 B4PX67 A0A1W3JUC2 A0A232F7G8 A0A182MCH4 H2ZJ45 A0A1A9ZYI2 A0A1S3JUL5 A0A1S3KBE9 V4BGM1 A0A182NFU6 A0A182SXL3 A0A2P8XPV8 W4Y2G3 A0A1B0G4P1 A0A0M4EV90 Q32TF6 C3ZDQ1 Q28X04 A0A3Q3LX26 B4MAX2 B4H4U8 B4JYU2 B4L5F9 W5M1L2 A0A3Q3LX21 A0A0R3U2Y8 A0A3P9Q8S6 A0A3B4UJD2 A0A3B4UIV3 A0A210QEN1 A0A3B4EF34 A0A3B5B1C8 A0A2B4SEE0 A0A2G8LEF1 B3MGS7 H3AR71 A7SUE4 A0A0X3PC41 A0A1Y1KSM3 G4V5T5 A0A095APE5 W5K4R5 A0A3Q0KSQ7 A0A3P9AL42 B5DEU8 A0A3P9AL37
PDB
2Z13
E-value=9.04731e-24,
Score=276
Ontologies
GO
PANTHER
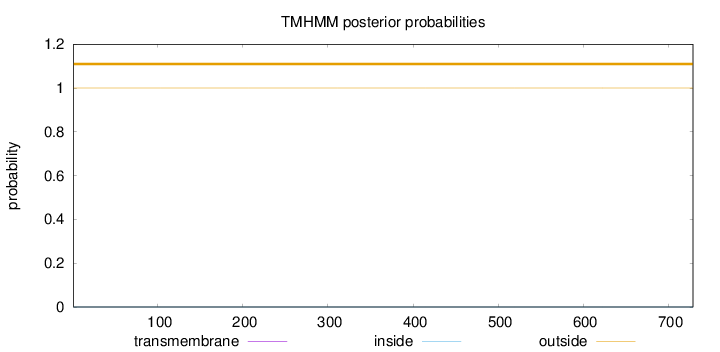
Topology
Length:
729
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00061
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00008
outside
1 - 729
Population Genetic Test Statistics
Pi
223.95255
Theta
167.176889
Tajima's D
1.034228
CLR
0
CSRT
0.670266486675666
Interpretation
Uncertain
