Gene
KWMTBOMO04576
Pre Gene Modal
BGIBMGA005478
Annotation
Dual_oxidase_[Papilio_machaon]
Full name
Dual oxidase
Location in the cell
PlasmaMembrane Reliability : 3.515
Sequence
CDS
ATGATGGCGTTCAAATGTTTAACGCCATTACATCGACTGTTATCAATAACAATCATCGCCGCCGTTCTCTCTTCGTGCTACTGCGACCCAGAGGTTTACTACGAGAAACAGCGGTACGATGGATGGTTTAATAACAGAGCGTATCCAGATTGGGGATCGGTTGGCAGTCGATTGAATCGTAAAACTCCAGCGTCATACGCTGACGGCGTTTATATGATGGCCGGTCCTGAGAGACCTGGTGCCAGGACCCTTTCGAAGATCTTCATGAGGGGTCAGGATGGACTGCCCTCACTAACGAATAGAACAGCGCTACTTGCATTCTTCGGGCAAGTGGTGACGGGCGAGATTGTAATGGCCTCAGAGTCCGGATGTCCTATCGAGCATCATCGTATACCGGTCGATAAATGCGATCATATGTACGACTCGGAATGTCAAGGTGCCAAGTATATGCCCTTTTTGCGAGCTGCGTACGACCGCAATACTGGACAGAGTCCCAACAGCCCCCGGGAACAAATAAATCAGATGACTTCGTGGATTGACGGGAGCTTCGTGTACAGCACGAGTGAGGCATGGGTGAATGCAATGAGGTCATTTCAGAACGGAAGTTTCGCCAGCGACGGAGGGATGCCACTACGGAACACAAAGAGGGTACCGCTTTTCAACAACCCCGTGCCGCATTACATGAGGATGCTTAGCCCAGAAAGGCTATTTTTACTAGGAGATCCACGTACCAATCAAAATCCAGCAATGGTGAGCTTCGGCATACTGCTGTTCCGCTGGCACAACGTCGTCGCAGCCCGAGTACACAAACAGCATCCGGACTGGAGCGACGAACAACTGTTCCAGAGAGCGCGGAGGATTGTCATAGCCAGTTTGCAAAACATCATTTTGTATGAATACGTGCCAGCGTTCCTCGGAGCAGCGCTTCCGCCTTACGAAGGTTACAAGCCGGAAGTAGCTCCGGGAGTGACCCACGAGTTCGCCGTAGCAGCATTTAGATTCGGCCATACATTAGTGCCACCAGCTATTCTACTTAGAAATAGGAAATGCGGTTACGGCAGAGCGCCGGGAGGACATGACGCTATTAGATTGTGTCAGACATGGTGGGACGGAAATGACGTTATGTCTCAAGTTCCACTAGAAGAAGTTTTGATGGGGATGGCCTCACAACTATCCGAGAGAGAGGACGCTCTACTGTGTTCCGATGTACGAGATAATTTGTTCGGTCCCATGGAATTTTCTAGACGAGACCTTGGCGCTTTGAATATAATGAGAGGAAGGGACAATGGTGTTCCAGATTATAATACAGTTAGGGAATACTTCGGCCTACGAAAAATAAAGACATTCAACGAAATAAATCCTGATCTGTTCGAGAAAAATCCTGATCTATTACAAAAATTTATCCAAATTTACAACGGAGACGTCGATAATATAGATGTGTATATCGGTGGGATGCTGGAGTCTAATGGACATCCGGGTGAATTGTTTAGAGCCATTATAATAGATCAATTTACAAGAATACGAGATTCTGATCGATTTTGGTTCGAAAATGACAAGAACGGTATATTCACTCCTCGAGATATAGAAGAGCTTCGTCGTATAACTCTGTGGGATATAATAGTGAACAGTACAACAATAGGTGCCAAAGATATACAAAGGGACGTGTTCCATTGGCGAGCGGGTGATCCGTGTCCACAACCATACCAACTGAACGCTTCCAAGCTTCCACCCTGCAAATATTTGAAGGGCTACGATTATTTTGAGGGAAACGAATTCGCCTACATCTACATCTGTTTGATATTAGTGTTCGTTCCCATATTATGCGCCGGAGCTGGTTACGGGGTTGTGAAGCTACAGAACAGTCGCAGACGGCGTTTGAAGATTCAGCAAGAACACCTTAAAAATGCTCAATGTAAAGGTTCAGTCGATAAAATGGTCTGTCGAGAATGGGTCCACGCGTCACACAAGAGACTTGTAAAGCTACGGCTTGGTCCAGAATCAGCCTTGCACGTGACAGATCGTAAGGGAGAAAAACTACGGACGCTGCCGTTGGATCACACTGATCAACTTACCGTAGTTGAAAGTCAGGAAGGTCGGAACAACAAACGTCCCCTCGTCCTCGTACGAGTCCCTCGGGAACATGACTTAGTGCTTGAAATGGACTCCATCGCATCTCGGCGCAAGTTTTTAGTCAAACTCGACACTTTCCTGGCGCAGCACAAGAAAGCGTTGAATCTTACTCAGGGACCGCGTGAACAAATTTTGGCTACAGCTGAAACAAGGGAGCGTCGACAACGCAAGTTAGAACATTTCTTCAGAGAAGCCTACGCTATAACATTCGGCCTCGCGCCTGGAGAGAAGCGGAGGAGGTCTGAAGATGCCGATCCTGAGTCAATAGTTATGAGAACGTCACTATCGAAAAGTGAATTTGCTAGTGCACTTGGTATGAAAGGAGACGCAGTCTTCGTCAAAAAAATGTTCAATATCGTCGACAAAGATGGCGACGGCAGGATTTCTTTTCAGGAGTTCCTTGATACTGTAGTTCTATTCTCTCGAGGAGCTACTGAGGATAAGCTTCGTATAATATTTGATATGTGCGACAACGATAGAAACGGTGTTATCGATAAAGGCGAACTGAGCGAGATGCTGAGGTCGCTCGTGGAAATAGCAAGAACTACCTCACTGAGAGATGAACATGTCACTGAGCTGATTGACGGAATGTTTTCTGATGCCGGACTTCAACACAAAGACCATTTGACCTACTCCGATTTCAAGCTGATGATGAAAGAATACAAAGGAGAGTTCGTGGCCATAGGTCTGGATTGTAAAGGCGCTAAGCAAAATTTCCTCGACACATCAACGAACGTTGCAAGAATGACCAGCTTCCATATTGAGCCTTCGATGGAACAGACCAGGCACTGGCTACTATTAAAATGGGATACCTTAACAACATTCTTGGAAGAAAACAGGCAAAATATATTTTATCTTTTCATTTTCTACGTTGTGACCATAGGCCTGTTTGTAGAAAGGTTCGCCCACTATTCGTTTATGTCGGAACATCTAGACTTGCGTCATATTATGGGAGTCGGTATCGCCATCACCAGAGGATCGGCCGCCTCGCTATCTTTCTGCTATAGTCTACTGTTATTGACGATGTCCAGGAATCTTTTAACGAAATTGAAGGAATTCAGTATCCAACAATACATACCTCTGGACTCGAGCATCCAGTTTCACAAGATCGTGGCCTGTACGGCTCTATTCTTTTCCTTGCTGCACACCGCCGGTCATATGGTCAACTTCTACCACGTGTCCACACAGCCCGTCGAGAACCTTCGATGTTTGACCAAAGAAGTACATTTCACATCCGACTTCAGGCCCGGCATTACCTACTGGGTATTCCAGACTGTTACTGGTGTAAGTGGGGTGCTGTTATTCGTGATCATGTGTATAATATTCGTATTTGCACATCCCGTGATCCGTAAACGCGCGTATCCATGGTTTTGGCGCGCTCACTCGCTGCACGTCGCGCTCTACGCGCTGTGCTTAGTCCACGGACTCGCTCGCCTTACCGGAGCCCCTAGGTTCTGGATATTCTTCCTTGGACCGGCTATCATATACACTCTGGATAAGGTGGTGTCCTTGAGAACGGAGTACCTTGCTTTAGACGTTTTAGAAACGGAGATGTTACCATCAGACGTTATAAAGATTAAGTTCTACCGACCCCCTAACCTAAAATATCTATCAGGTCAATGGGTTCGTTTGTCGTGTACAGCGTTTAAGAAGGAGGAGTTCCATTCGTTTACACTCACATCAGCACCGCACGAGAACTTCCTCTCGTGTCACATCAAAGCACAAGGACCGTGGACTTGGAAACTAAGAAACTACTTCGACCCTTGCAATTATAATACAGACGATCATCCTAAAATCAGAATACAAGGTCCGTTCGGTGGCGGGAACCAGGACTGGTACAAGTTCGAGGTGGCCGTGATGGTGGGCGGAGGCATCGGCGTCACTCCCTACGCCTCCATACTTAACGACCTCGTATTCGGAACATCCACGAACAGATATTCTGGGGTTGCTTGTAAAAAGGTATATTTTCTATGGATATGTCCATCGCACAAACACTTCGAATGGTTCATAGACGTTCTCCGCGACGTCGAGCGGAAAGACGTGACTAACGTACTGGAAATCCACATCTTCATAACGCAATTCTTCCACAAGTTCGATCTGAGGACGACCATGCTGTACATATGCGAGAATCATTTCCAACGTCTGTCTCGGACGTCGATGTTCACGGGACTGAAGGCGGTCAACCACTTCGGCCGACCAGACATGTCCTCCTTCCTCAAGTTCGTACAGAAGAAACATAGTTACGTGTCAAAGATCGGTGTGTTTTCGTGTGGGCCCCGACCACTCACCAAGTCGGTGATGTCAGCTTGCGAAGAAGTAAACAGGACCAGGCGTCTGCCTTACTTCATACATCACTTTGAAAACTTTGGATAA
Protein
MMAFKCLTPLHRLLSITIIAAVLSSCYCDPEVYYEKQRYDGWFNNRAYPDWGSVGSRLNRKTPASYADGVYMMAGPERPGARTLSKIFMRGQDGLPSLTNRTALLAFFGQVVTGEIVMASESGCPIEHHRIPVDKCDHMYDSECQGAKYMPFLRAAYDRNTGQSPNSPREQINQMTSWIDGSFVYSTSEAWVNAMRSFQNGSFASDGGMPLRNTKRVPLFNNPVPHYMRMLSPERLFLLGDPRTNQNPAMVSFGILLFRWHNVVAARVHKQHPDWSDEQLFQRARRIVIASLQNIILYEYVPAFLGAALPPYEGYKPEVAPGVTHEFAVAAFRFGHTLVPPAILLRNRKCGYGRAPGGHDAIRLCQTWWDGNDVMSQVPLEEVLMGMASQLSEREDALLCSDVRDNLFGPMEFSRRDLGALNIMRGRDNGVPDYNTVREYFGLRKIKTFNEINPDLFEKNPDLLQKFIQIYNGDVDNIDVYIGGMLESNGHPGELFRAIIIDQFTRIRDSDRFWFENDKNGIFTPRDIEELRRITLWDIIVNSTTIGAKDIQRDVFHWRAGDPCPQPYQLNASKLPPCKYLKGYDYFEGNEFAYIYICLILVFVPILCAGAGYGVVKLQNSRRRRLKIQQEHLKNAQCKGSVDKMVCREWVHASHKRLVKLRLGPESALHVTDRKGEKLRTLPLDHTDQLTVVESQEGRNNKRPLVLVRVPREHDLVLEMDSIASRRKFLVKLDTFLAQHKKALNLTQGPREQILATAETRERRQRKLEHFFREAYAITFGLAPGEKRRRSEDADPESIVMRTSLSKSEFASALGMKGDAVFVKKMFNIVDKDGDGRISFQEFLDTVVLFSRGATEDKLRIIFDMCDNDRNGVIDKGELSEMLRSLVEIARTTSLRDEHVTELIDGMFSDAGLQHKDHLTYSDFKLMMKEYKGEFVAIGLDCKGAKQNFLDTSTNVARMTSFHIEPSMEQTRHWLLLKWDTLTTFLEENRQNIFYLFIFYVVTIGLFVERFAHYSFMSEHLDLRHIMGVGIAITRGSAASLSFCYSLLLLTMSRNLLTKLKEFSIQQYIPLDSSIQFHKIVACTALFFSLLHTAGHMVNFYHVSTQPVENLRCLTKEVHFTSDFRPGITYWVFQTVTGVSGVLLFVIMCIIFVFAHPVIRKRAYPWFWRAHSLHVALYALCLVHGLARLTGAPRFWIFFLGPAIIYTLDKVVSLRTEYLALDVLETEMLPSDVIKIKFYRPPNLKYLSGQWVRLSCTAFKKEEFHSFTLTSAPHENFLSCHIKAQGPWTWKLRNYFDPCNYNTDDHPKIRIQGPFGGGNQDWYKFEVAVMVGGGIGVTPYASILNDLVFGTSTNRYSGVACKKVYFLWICPSHKHFEWFIDVLRDVERKDVTNVLEIHIFITQFFHKFDLRTTMLYICENHFQRLSRTSMFTGLKAVNHFGRPDMSSFLKFVQKKHSYVSKIGVFSCGPRPLTKSVMSACEEVNRTRRLPYFIHHFENFG
Summary
Description
Plays a role in innate immunity limiting microbial proliferation in the gut (PubMed:16272120, PubMed:25639794). Acts downstream of a hh-signaling pathway to induce the production of reactive oxygen species (ROS) in response to intestinal bacterial infection (PubMed:25639794). May generate antimicrobial oxidative burst through its peroxidase-like domain (PubMed:16272120).
Catalytic Activity
H(+) + NADH + O2 = H2O2 + NAD(+)
H(+) + NADPH + O2 = H2O2 + NADP(+)
H(+) + NADPH + O2 = H2O2 + NADP(+)
Similarity
In the N-terminal section; belongs to the peroxidase family.
Keywords
Calcium
Complete proteome
FAD
Flavoprotein
Glycoprotein
Hydrogen peroxide
Membrane
Metal-binding
NADP
Oxidoreductase
Peroxidase
Phosphoprotein
Reference proteome
Repeat
Transmembrane
Transmembrane helix
Feature
chain Dual oxidase
Uniprot
R4IPZ2
H9J7I6
A0A194QSN3
A0A3S2LF46
A0A194PLQ5
A0A212ES06
+ More
A0A2H1V4L3 A0A2M4CTD8 A0A1S4H2M1 A0A182HRA5 A0A182MST9 A0A2J7RBS7 Q7Q147 A0A182XGX2 A0A084VSU8 A0A182F181 A0A182Y6B3 A0A182Q2T7 A0A182JX33 A0A1Y0K3W4 A0A182IYI7 W4VRJ7 B0W719 A0A1Q3EVH1 A0A1I8PXE3 A0A1I8PXC7 A0A1I8NGL1 M9PCF1 Q9VQH2 A0A0J9QUP3 D6WH98 Q171Q3 A0A1W4V601 Q29KF2 D0Z752 A0A182T0U4 A0A182RWS0 A0A182W8S4 B3NA67 B4N013 A0A034V5W5 A0A182NP34 B4JZF3 B4NX88 A0A0M4E2I1 B4LUX5 A0A0K8VLK7 A0A0K0Y0T6 A0A0A1WMN8 W5J7V8 A0A3B0JNF8 W8AIH8 A0A1W4WU34 A0A3B0JPT7 A0A1B0D6K5 B4KF94 A0A0L0CQ69 A0A1A9ZHR8 A0A1B6CWK3 A0A182PJL3 A0A336LKU5 A0A1A9YJJ7 A0A1B0BP54 B4Q8L1 A0A1B6MRT5 A0A1A9X483 B4HBY1 A0A3L8E2B0 B3MLA0 A0A026WLE4 A0A088AIX9 A0A0N0BC71 A0A1J1I5A5 A0A195D3J9 A0A158NQQ1 A7E3M3 A0A182V5M4 A0A182KZ29 A0A182TTX1 F4WBV9 E1ZXB1 A0A226EPH4 A0A151XB50 A0A195DI90 A0A195FSQ1 A0A195BDA0 T1HLF9 A0A2A3E4D0 A0A1A9UIU5 A0A146KVX5 A0A0A9W1Z6 A0A2S2Q9Q5 I3RSJ6 E0VUS0 A0A0P5W941 E9HFQ7 A0A0A9VZ89 J9JPD3 K7IRZ2 A0A0P6IQJ2
A0A2H1V4L3 A0A2M4CTD8 A0A1S4H2M1 A0A182HRA5 A0A182MST9 A0A2J7RBS7 Q7Q147 A0A182XGX2 A0A084VSU8 A0A182F181 A0A182Y6B3 A0A182Q2T7 A0A182JX33 A0A1Y0K3W4 A0A182IYI7 W4VRJ7 B0W719 A0A1Q3EVH1 A0A1I8PXE3 A0A1I8PXC7 A0A1I8NGL1 M9PCF1 Q9VQH2 A0A0J9QUP3 D6WH98 Q171Q3 A0A1W4V601 Q29KF2 D0Z752 A0A182T0U4 A0A182RWS0 A0A182W8S4 B3NA67 B4N013 A0A034V5W5 A0A182NP34 B4JZF3 B4NX88 A0A0M4E2I1 B4LUX5 A0A0K8VLK7 A0A0K0Y0T6 A0A0A1WMN8 W5J7V8 A0A3B0JNF8 W8AIH8 A0A1W4WU34 A0A3B0JPT7 A0A1B0D6K5 B4KF94 A0A0L0CQ69 A0A1A9ZHR8 A0A1B6CWK3 A0A182PJL3 A0A336LKU5 A0A1A9YJJ7 A0A1B0BP54 B4Q8L1 A0A1B6MRT5 A0A1A9X483 B4HBY1 A0A3L8E2B0 B3MLA0 A0A026WLE4 A0A088AIX9 A0A0N0BC71 A0A1J1I5A5 A0A195D3J9 A0A158NQQ1 A7E3M3 A0A182V5M4 A0A182KZ29 A0A182TTX1 F4WBV9 E1ZXB1 A0A226EPH4 A0A151XB50 A0A195DI90 A0A195FSQ1 A0A195BDA0 T1HLF9 A0A2A3E4D0 A0A1A9UIU5 A0A146KVX5 A0A0A9W1Z6 A0A2S2Q9Q5 I3RSJ6 E0VUS0 A0A0P5W941 E9HFQ7 A0A0A9VZ89 J9JPD3 K7IRZ2 A0A0P6IQJ2
Pubmed
23936382
19121390
26354079
22118469
12364791
24438588
+ More
25244985 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 16272120 17372656 18327897 25639794 22936249 18362917 19820115 17510324 15632085 17994087 23185243 25348373 17550304 18057021 25830018 20920257 23761445 24495485 26108605 30249741 24508170 21347285 16381931 17612411 20966253 21719571 20798317 26823975 25401762 20566863 21292972 20075255
25244985 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 16272120 17372656 18327897 25639794 22936249 18362917 19820115 17510324 15632085 17994087 23185243 25348373 17550304 18057021 25830018 20920257 23761445 24495485 26108605 30249741 24508170 21347285 16381931 17612411 20966253 21719571 20798317 26823975 25401762 20566863 21292972 20075255
EMBL
JQ768349
AFV61649.1
BABH01013910
BABH01013911
BABH01013912
BABH01013913
+ More
BABH01013914 KQ461154 KPJ08497.1 RSAL01000163 RVE45365.1 KQ459599 KPI94366.1 AGBW02012896 OWR44265.1 ODYU01000617 SOQ35716.1 GGFL01004263 MBW68441.1 AAAB01008980 APCN01001661 APCN01001662 AXCM01001826 NEVH01005891 PNF38268.1 EAA13921.4 ATLV01016141 KE525057 KFB41042.1 AXCN02001965 KY386660 ARU79623.1 AXCP01008099 GANO01002274 JAB57597.1 DS231851 EDS37293.1 GFDL01015736 JAV19309.1 AE014134 AGB92505.1 BT015247 CM002910 KMY87751.1 KQ971330 EFA01258.2 CH477448 EAT40728.2 CH379061 EAL33223.2 KRT04822.1 BT100299 ACZ06885.1 CH954177 EDV57530.2 CH963920 EDW77948.1 GAKP01021421 GAKP01021420 JAC37532.1 CH916379 EDV94075.1 CM000157 EDW87445.2 CP012523 ALC39816.1 CH940649 EDW63224.2 KRF80974.1 GDHF01012548 JAI39766.1 KP824741 AKS43593.1 GBXI01014165 GBXI01009467 GBXI01002107 JAD00127.1 JAD04825.1 JAD12185.1 ADMH02002110 ETN58940.1 OUUW01000006 SPP82413.1 GAMC01021098 GAMC01021097 GAMC01021096 GAMC01021095 JAB85457.1 SPP82412.1 AJVK01003690 AJVK01003691 AJVK01003692 CH933807 EDW11994.2 JRES01000175 KNC33589.1 GEDC01019500 JAS17798.1 UFQT01000040 SSX18662.1 JXJN01017817 CM000361 EDX03528.1 GEBQ01001336 JAT38641.1 CH479272 EDW39992.1 QOIP01000001 RLU26860.1 CH902620 EDV30689.2 KK107154 EZA56872.1 KQ435922 KOX68568.1 CVRI01000038 CRK94094.1 KQ976885 KYN07485.1 ADTU01023460 BR000285 FAA00352.1 GL888066 EGI68387.1 GL435030 EFN74201.1 LNIX01000003 OXA58721.1 KQ982335 KYQ57577.1 KQ980824 KYN12552.1 KQ981280 KYN43473.1 KQ976511 KYM82538.1 ACPB03010969 ACPB03010970 ACPB03010971 ACPB03010972 KZ288405 PBC26142.1 GDHC01019283 JAP99345.1 GBHO01043131 JAG00473.1 GGMS01005077 MBY74280.1 JQ266749 AFK29281.1 DS235793 EEB17126.1 GDIP01089476 LRGB01000086 JAM14239.1 KZS20913.1 GL732637 EFX69361.1 GBHO01043128 JAG00476.1 ABLF02030921 GDIQ01007363 JAN87374.1
BABH01013914 KQ461154 KPJ08497.1 RSAL01000163 RVE45365.1 KQ459599 KPI94366.1 AGBW02012896 OWR44265.1 ODYU01000617 SOQ35716.1 GGFL01004263 MBW68441.1 AAAB01008980 APCN01001661 APCN01001662 AXCM01001826 NEVH01005891 PNF38268.1 EAA13921.4 ATLV01016141 KE525057 KFB41042.1 AXCN02001965 KY386660 ARU79623.1 AXCP01008099 GANO01002274 JAB57597.1 DS231851 EDS37293.1 GFDL01015736 JAV19309.1 AE014134 AGB92505.1 BT015247 CM002910 KMY87751.1 KQ971330 EFA01258.2 CH477448 EAT40728.2 CH379061 EAL33223.2 KRT04822.1 BT100299 ACZ06885.1 CH954177 EDV57530.2 CH963920 EDW77948.1 GAKP01021421 GAKP01021420 JAC37532.1 CH916379 EDV94075.1 CM000157 EDW87445.2 CP012523 ALC39816.1 CH940649 EDW63224.2 KRF80974.1 GDHF01012548 JAI39766.1 KP824741 AKS43593.1 GBXI01014165 GBXI01009467 GBXI01002107 JAD00127.1 JAD04825.1 JAD12185.1 ADMH02002110 ETN58940.1 OUUW01000006 SPP82413.1 GAMC01021098 GAMC01021097 GAMC01021096 GAMC01021095 JAB85457.1 SPP82412.1 AJVK01003690 AJVK01003691 AJVK01003692 CH933807 EDW11994.2 JRES01000175 KNC33589.1 GEDC01019500 JAS17798.1 UFQT01000040 SSX18662.1 JXJN01017817 CM000361 EDX03528.1 GEBQ01001336 JAT38641.1 CH479272 EDW39992.1 QOIP01000001 RLU26860.1 CH902620 EDV30689.2 KK107154 EZA56872.1 KQ435922 KOX68568.1 CVRI01000038 CRK94094.1 KQ976885 KYN07485.1 ADTU01023460 BR000285 FAA00352.1 GL888066 EGI68387.1 GL435030 EFN74201.1 LNIX01000003 OXA58721.1 KQ982335 KYQ57577.1 KQ980824 KYN12552.1 KQ981280 KYN43473.1 KQ976511 KYM82538.1 ACPB03010969 ACPB03010970 ACPB03010971 ACPB03010972 KZ288405 PBC26142.1 GDHC01019283 JAP99345.1 GBHO01043131 JAG00473.1 GGMS01005077 MBY74280.1 JQ266749 AFK29281.1 DS235793 EEB17126.1 GDIP01089476 LRGB01000086 JAM14239.1 KZS20913.1 GL732637 EFX69361.1 GBHO01043128 JAG00476.1 ABLF02030921 GDIQ01007363 JAN87374.1
Proteomes
UP000005204
UP000053240
UP000283053
UP000053268
UP000007151
UP000075840
+ More
UP000075883 UP000235965 UP000007062 UP000076407 UP000030765 UP000069272 UP000076408 UP000075886 UP000075881 UP000075880 UP000002320 UP000095300 UP000095301 UP000000803 UP000007266 UP000008820 UP000192221 UP000001819 UP000075901 UP000075900 UP000075920 UP000008711 UP000007798 UP000075884 UP000001070 UP000002282 UP000092553 UP000008792 UP000000673 UP000268350 UP000192223 UP000092462 UP000009192 UP000037069 UP000092445 UP000075885 UP000092443 UP000092460 UP000000304 UP000091820 UP000008744 UP000279307 UP000007801 UP000053097 UP000005203 UP000053105 UP000183832 UP000078542 UP000005205 UP000075903 UP000075882 UP000075902 UP000007755 UP000000311 UP000198287 UP000075809 UP000078492 UP000078541 UP000078540 UP000015103 UP000242457 UP000078200 UP000009046 UP000076858 UP000000305 UP000007819 UP000002358
UP000075883 UP000235965 UP000007062 UP000076407 UP000030765 UP000069272 UP000076408 UP000075886 UP000075881 UP000075880 UP000002320 UP000095300 UP000095301 UP000000803 UP000007266 UP000008820 UP000192221 UP000001819 UP000075901 UP000075900 UP000075920 UP000008711 UP000007798 UP000075884 UP000001070 UP000002282 UP000092553 UP000008792 UP000000673 UP000268350 UP000192223 UP000092462 UP000009192 UP000037069 UP000092445 UP000075885 UP000092443 UP000092460 UP000000304 UP000091820 UP000008744 UP000279307 UP000007801 UP000053097 UP000005203 UP000053105 UP000183832 UP000078542 UP000005205 UP000075903 UP000075882 UP000075902 UP000007755 UP000000311 UP000198287 UP000075809 UP000078492 UP000078541 UP000078540 UP000015103 UP000242457 UP000078200 UP000009046 UP000076858 UP000000305 UP000007819 UP000002358
Pfam
Interpro
IPR037120
Haem_peroxidase_sf_animal
+ More
IPR019791 Haem_peroxidase_animal
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
IPR039261 FNR_nucleotide-bd
IPR013130 Fe3_Rdtase_TM_dom
IPR034821 DUOX_peroxidase
IPR017927 FAD-bd_FR_type
IPR013112 FAD-bd_8
IPR017938 Riboflavin_synthase-like_b-brl
IPR013121 Fe_red_NAD-bd_6
IPR010255 Haem_peroxidase_sf
IPR018247 EF_Hand_1_Ca_BS
IPR029595 DUOX1/Duox
IPR019791 Haem_peroxidase_animal
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
IPR039261 FNR_nucleotide-bd
IPR013130 Fe3_Rdtase_TM_dom
IPR034821 DUOX_peroxidase
IPR017927 FAD-bd_FR_type
IPR013112 FAD-bd_8
IPR017938 Riboflavin_synthase-like_b-brl
IPR013121 Fe_red_NAD-bd_6
IPR010255 Haem_peroxidase_sf
IPR018247 EF_Hand_1_Ca_BS
IPR029595 DUOX1/Duox
Gene 3D
ProteinModelPortal
R4IPZ2
H9J7I6
A0A194QSN3
A0A3S2LF46
A0A194PLQ5
A0A212ES06
+ More
A0A2H1V4L3 A0A2M4CTD8 A0A1S4H2M1 A0A182HRA5 A0A182MST9 A0A2J7RBS7 Q7Q147 A0A182XGX2 A0A084VSU8 A0A182F181 A0A182Y6B3 A0A182Q2T7 A0A182JX33 A0A1Y0K3W4 A0A182IYI7 W4VRJ7 B0W719 A0A1Q3EVH1 A0A1I8PXE3 A0A1I8PXC7 A0A1I8NGL1 M9PCF1 Q9VQH2 A0A0J9QUP3 D6WH98 Q171Q3 A0A1W4V601 Q29KF2 D0Z752 A0A182T0U4 A0A182RWS0 A0A182W8S4 B3NA67 B4N013 A0A034V5W5 A0A182NP34 B4JZF3 B4NX88 A0A0M4E2I1 B4LUX5 A0A0K8VLK7 A0A0K0Y0T6 A0A0A1WMN8 W5J7V8 A0A3B0JNF8 W8AIH8 A0A1W4WU34 A0A3B0JPT7 A0A1B0D6K5 B4KF94 A0A0L0CQ69 A0A1A9ZHR8 A0A1B6CWK3 A0A182PJL3 A0A336LKU5 A0A1A9YJJ7 A0A1B0BP54 B4Q8L1 A0A1B6MRT5 A0A1A9X483 B4HBY1 A0A3L8E2B0 B3MLA0 A0A026WLE4 A0A088AIX9 A0A0N0BC71 A0A1J1I5A5 A0A195D3J9 A0A158NQQ1 A7E3M3 A0A182V5M4 A0A182KZ29 A0A182TTX1 F4WBV9 E1ZXB1 A0A226EPH4 A0A151XB50 A0A195DI90 A0A195FSQ1 A0A195BDA0 T1HLF9 A0A2A3E4D0 A0A1A9UIU5 A0A146KVX5 A0A0A9W1Z6 A0A2S2Q9Q5 I3RSJ6 E0VUS0 A0A0P5W941 E9HFQ7 A0A0A9VZ89 J9JPD3 K7IRZ2 A0A0P6IQJ2
A0A2H1V4L3 A0A2M4CTD8 A0A1S4H2M1 A0A182HRA5 A0A182MST9 A0A2J7RBS7 Q7Q147 A0A182XGX2 A0A084VSU8 A0A182F181 A0A182Y6B3 A0A182Q2T7 A0A182JX33 A0A1Y0K3W4 A0A182IYI7 W4VRJ7 B0W719 A0A1Q3EVH1 A0A1I8PXE3 A0A1I8PXC7 A0A1I8NGL1 M9PCF1 Q9VQH2 A0A0J9QUP3 D6WH98 Q171Q3 A0A1W4V601 Q29KF2 D0Z752 A0A182T0U4 A0A182RWS0 A0A182W8S4 B3NA67 B4N013 A0A034V5W5 A0A182NP34 B4JZF3 B4NX88 A0A0M4E2I1 B4LUX5 A0A0K8VLK7 A0A0K0Y0T6 A0A0A1WMN8 W5J7V8 A0A3B0JNF8 W8AIH8 A0A1W4WU34 A0A3B0JPT7 A0A1B0D6K5 B4KF94 A0A0L0CQ69 A0A1A9ZHR8 A0A1B6CWK3 A0A182PJL3 A0A336LKU5 A0A1A9YJJ7 A0A1B0BP54 B4Q8L1 A0A1B6MRT5 A0A1A9X483 B4HBY1 A0A3L8E2B0 B3MLA0 A0A026WLE4 A0A088AIX9 A0A0N0BC71 A0A1J1I5A5 A0A195D3J9 A0A158NQQ1 A7E3M3 A0A182V5M4 A0A182KZ29 A0A182TTX1 F4WBV9 E1ZXB1 A0A226EPH4 A0A151XB50 A0A195DI90 A0A195FSQ1 A0A195BDA0 T1HLF9 A0A2A3E4D0 A0A1A9UIU5 A0A146KVX5 A0A0A9W1Z6 A0A2S2Q9Q5 I3RSJ6 E0VUS0 A0A0P5W941 E9HFQ7 A0A0A9VZ89 J9JPD3 K7IRZ2 A0A0P6IQJ2
PDB
6ERC
E-value=3.42581e-54,
Score=541
Ontologies
PATHWAY
GO
GO:0050665
GO:0004601
GO:0016021
GO:0016174
GO:0020037
GO:0005509
GO:0006979
GO:0016491
GO:0005886
GO:0043020
GO:0006952
GO:0019221
GO:0055114
GO:0042554
GO:0042335
GO:0051591
GO:0016175
GO:0012505
GO:0043066
GO:0042742
GO:0042744
GO:0008365
GO:0072593
GO:0048085
GO:0035220
GO:0007476
GO:0002385
GO:0009611
GO:0007165
GO:0015031
GO:0016020
GO:0016742
GO:0003824
PANTHER
Topology
Subcellular location
Membrane
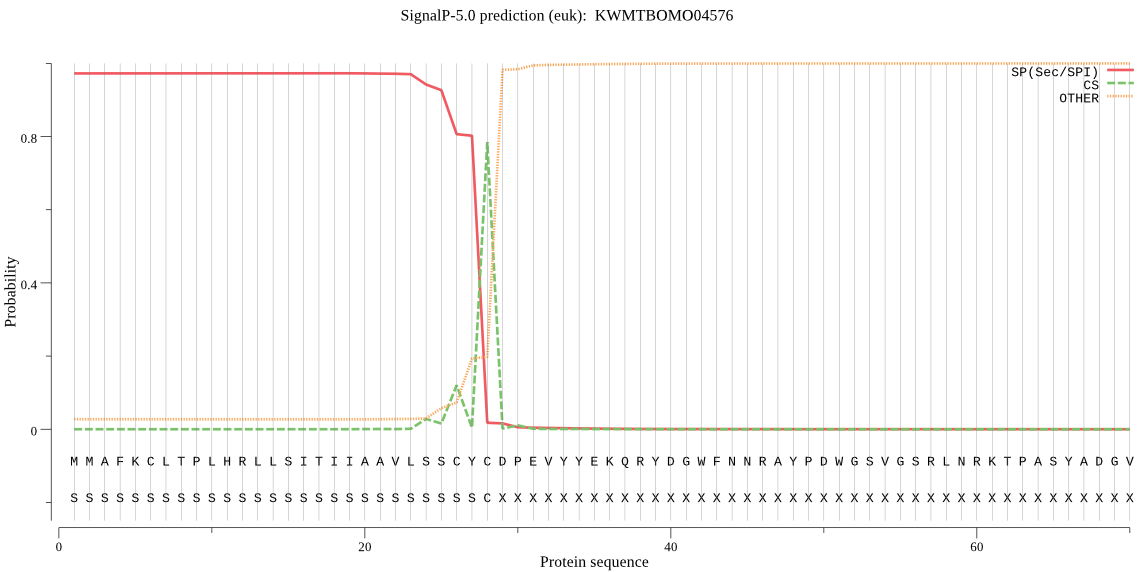
SignalP
Position: 1 - 28,
Likelihood: 0.972961
Length:
1500
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
136.75384
Exp number, first 60 AAs:
0.67935
Total prob of N-in:
0.03132
outside
1 - 592
TMhelix
593 - 615
inside
616 - 992
TMhelix
993 - 1012
outside
1013 - 1026
TMhelix
1027 - 1049
inside
1050 - 1079
TMhelix
1080 - 1102
outside
1103 - 1131
TMhelix
1132 - 1154
inside
1155 - 1174
TMhelix
1175 - 1197
outside
1198 - 1500

Population Genetic Test Statistics
Pi
282.630374
Theta
198.438871
Tajima's D
1.010968
CLR
0.330731
CSRT
0.661566921653917
Interpretation
Uncertain
