Gene
KWMTBOMO04573 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005477
Annotation
integrin_alpha-PS1_precursor_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.126 PlasmaMembrane Reliability : 1.758
Sequence
CDS
ATGTACCTGCTATCCGTTAATTGCACCATGTTACTTGTTGGAGCTCCGTTAGATCAAAATCTACAACCGAACACAACAAAGTCTGGCGCTTTGTGGAAATGCAGTCTCAGCCCAGACACGAACGACTGCGAGCAGATTATTACAGACGGGAAACGAACACTGGATTCTGATCATCTATCGGGTCCTTATTCAGACGAGATCAAGGAAGGCCAATGGCTCGGCGTGACTGTTCGAAGTCAGGGTCCTGGCAAAAAGGCTGTAGTTTGTGCTCACAGGTACATTCGAAAGATTGGACAACTGCAATTCGGCCAAGGACTGTGTTACACTTTCAAAAACACTCTACAACTACACGAAATCACTGAACCTTGTAGAGGAAGACCTATTACTAGAGAACACGAAGAATATGGTTTCTGTCAAGTAGGAACAAGCAGTACTCTATTGGATGATGAAACATTAATCCTCGGCAGTCCGGGACCGTATGCCTGGCGAGGAACAATATTCGCACAAGATATGACCGACGAAATGTTTGCCCGAGATAATGTGGTCTATAGTGCACCTGTCACTGATGGTGCTAGTCCTGTCGAGAAATACAGCTACCTGGGAATGTCAGTAGCAGGTGGAATGTTTTTTGGAAACAAGTCCTCTTACGTAGCCGGTGCACCCCGGGCCCAAGGGACTGGTCAAGTTGTAATTTTCAACAAAAAAGACACCGCTGATTGGGGCAGAACGGACTTAGATATTCTTCGTTTTACACTCATTCTGAAAGGGGAACAGTTTGGGTCCAGCTTTGGATATGAAGTGGCCGTGGCTGATGTTAATGGAGATGGATTGCCGGATCTTCTAGTCGGTGCACCATTTTATTTTTCACGAGATGCCGGTGGTGCAGTGTACTTATATCTTAATAAAAATCAAAACTTAACAAAAAAGTTCGATTTAAAGTTGACTGGAAAACAAGAATCACAGTTTGGTATTTCCATTGCTAATGCATCAGACTTAAACAAAGATGGTTGTCAAGATATAGCAATCGGTAGTCCTTACGAAGGGAATGGAGTTGTGTATATTTATATGGGTAACCGTAGAACTGGTTTGAATTCAATACCAGATCAGGTTATTTCAGCTGAAAGCTTACCAATCGTATTGAAGACATTTGGCTACTCGCTTTCAGGGGGTATTGATTTAGATGCTAATGGATATCCTGATTTGTTAGTTGGCGCGTATGAGAATAGCAGCGTCGCTTTAATACGAACGAGACCGATCATTGACATAAAGACGTCAATACGACCATCGAATACTATAATTAATATCGACCCGACGAAGCAGGGCTGTGAAAGAGATCCATCCTCAAAGTATACATGTTTCCACTTTCAAGCCTGCTGTACAATCGAATCTTTGGTGAAATCCACAAAAACGAATGCTCTAAAGTTGAATTATGTAATTGAAGCAGAAACTTTCCTTGGCGGGAAAAGATATTCTAGGGTGTTCTTGGATACAGAAAAGACGAACATAGTTAAAAAGTCAATTGTTCTGGACAAAGAAATCGAAGATTGCAGAGAACACATTGTTTACATAAAAAACAATACAAGAGATATTCAAACCCCAATAAAGTTCCAAGTGACATATAAATTGGAGAATGTCGAGCCGCAATATTCTCCCTCTGGCACTTTGCCCAACATCGATGATTACCCTGTTCTCAATGCGACAGCGATGACAACCTTCAGCGCTCACTTCTTGAACGACTGCGGGACTGATGCGATTTGTGTTAGTAATCTAATCGTCAATCCTGTTTTACAATTACCAAAAACGTCAGATGGCAAGTCGTACGCGCTCACATTAGGCCAAGAGGAAGAAATTAAGCTGTCAATAACTGTAGACAATTATGGGGAGTCAGCATACGAGAGCCAACTTTTCGTACAACACCATTCAAGTTTACATTACATCGCTGCAAATATAAGTGACAAGAACGTGATATGTACGAGTGTCAATAAAACGATAGTATCTTGTATGCTGGAGAATCCTTTCAAGAAGCAGCCGGAAAGTGCGGCTCCCATCACAATGAGGTTCGACGCAAGAGCACTTGAAGACAATGAGCCTTCAGTAGTGTTCACAGTTTGGGCTAATTCTACCTCCAAAGAATTGCACTCTGGGAAGAAGCCGGTTCAAGTCGAAGCCCTTGTAATTAAGAATGCTGAATTACTAATTAAAGGAGCTGCCCGACCAGAACAAGTTTTCTACGGAGGAGATATAAAAGGAGAATCTGCCATGACTTACTTCGATGACGTCGGTACAAGAGTGGTGCATACTTACCAGGTATTCAATGAAGGCCCTTGGCGCGTGTCTTCCGTGCAAGTTGTTATATTGTGGCCACATCAGGTAGCTTCTGAAAACGCACAGGGCAAATGGCTACTGTATCCCGAAGATGTTCCTACCGTCGACGGCGAAGCTGACCAGAGTAATGGTGAGTGCTTTATCACAGAAAAAGAGATTAACCCCTTAAACCTGACGTCCAGACCTGGAACATTTGAGCCTTATTTAGAAAACTTGGAGATAAATCCATTCATGAGTACTGGAAAGATCAGCGTGAACTCTTATGAAAAACAACAAAATCAAAGTAAAGTTGTAGAATTATCTACTGCCAAGGAAAATAAAGTGAGAAGAAAAAGAGATAGTGACGTAGTTAAAGCCGAAGCATATACGGACAAAGATGGGCAGAGAAAACATGTGGTTAATATGAGCTGCCAAAAGAAGACAGCGAAGTGCATACAATTTCAATGCGTGATCTATCGACTATCTCGGGCTCAGGCCACAACCATTACTGTGAAGGCTCGACTTTGGAACGCTACACTAGTAGAAGATTATCCCCGAATCAGCCATGTCAACATCGCTTCTACTGCGTTCATACATATACCAGATCACTACAACATCCATCAAAGCAAACAACACGATGATACCGCCACGGTTGAAACAGTCGCATATCCGGATTTGAAGATCAGCGAGCCCACAGAAGTTCCTCTTTGGGTCATCATTATATCAGTAATTATTGGACTAATCGTTCTGATACTCCTCATCTTAGCCCTTTGGAAACTGGGCTTCTTCAAGAGAAGTCGTCCTGACCCTACTCTCTCAGGAAACTTGGAGAAAAACCACCACGAATCCAGTCCGTTCATAGGAAGAGATAGAAATAGCATTCGATAG
Protein
MYLLSVNCTMLLVGAPLDQNLQPNTTKSGALWKCSLSPDTNDCEQIITDGKRTLDSDHLSGPYSDEIKEGQWLGVTVRSQGPGKKAVVCAHRYIRKIGQLQFGQGLCYTFKNTLQLHEITEPCRGRPITREHEEYGFCQVGTSSTLLDDETLILGSPGPYAWRGTIFAQDMTDEMFARDNVVYSAPVTDGASPVEKYSYLGMSVAGGMFFGNKSSYVAGAPRAQGTGQVVIFNKKDTADWGRTDLDILRFTLILKGEQFGSSFGYEVAVADVNGDGLPDLLVGAPFYFSRDAGGAVYLYLNKNQNLTKKFDLKLTGKQESQFGISIANASDLNKDGCQDIAIGSPYEGNGVVYIYMGNRRTGLNSIPDQVISAESLPIVLKTFGYSLSGGIDLDANGYPDLLVGAYENSSVALIRTRPIIDIKTSIRPSNTIINIDPTKQGCERDPSSKYTCFHFQACCTIESLVKSTKTNALKLNYVIEAETFLGGKRYSRVFLDTEKTNIVKKSIVLDKEIEDCREHIVYIKNNTRDIQTPIKFQVTYKLENVEPQYSPSGTLPNIDDYPVLNATAMTTFSAHFLNDCGTDAICVSNLIVNPVLQLPKTSDGKSYALTLGQEEEIKLSITVDNYGESAYESQLFVQHHSSLHYIAANISDKNVICTSVNKTIVSCMLENPFKKQPESAAPITMRFDARALEDNEPSVVFTVWANSTSKELHSGKKPVQVEALVIKNAELLIKGAARPEQVFYGGDIKGESAMTYFDDVGTRVVHTYQVFNEGPWRVSSVQVVILWPHQVASENAQGKWLLYPEDVPTVDGEADQSNGECFITEKEINPLNLTSRPGTFEPYLENLEINPFMSTGKISVNSYEKQQNQSKVVELSTAKENKVRRKRDSDVVKAEAYTDKDGQRKHVVNMSCQKKTAKCIQFQCVIYRLSRAQATTITVKARLWNATLVEDYPRISHVNIASTAFIHIPDHYNIHQSKQHDDTATVETVAYPDLKISEPTEVPLWVIIISVIIGLIVLILLILALWKLGFFKRSRPDPTLSGNLEKNHHESSPFIGRDRNSIR
Summary
Subunit
Heterodimer of an alpha and a beta subunit.
Similarity
Belongs to the integrin alpha chain family.
Uniprot
A0A076JVB3
H9J7I5
A0A212ES31
A0A194PNQ1
A0A2H1VCY1
A0A1E1WRC4
+ More
A0A2A4JVZ9 A0A194QU26 Q86G88 A0A1E1WLE5 A0A0L7LQM8 A0A067QUQ4 A0A1L8E400 A0A1L8E3C4 A0A1L8E3J4 A0A026VUY3 A0A1B0GKX3 A0A2A3ECL3 A0A3L8DE97 F4X5Q1 A0A195FQK3 A0A088ACX9 A0A154P056 A0A310STA2 A0A0P5N3H1 A0A0N8BB58 A0A0P6GVW7 A0A0P4ZC31 E9HAH4 A0A0P5C057 A0A0P4XX18 A0A0N8CM66 A0A0N8BWE9 A0A195BWJ3 A0A0N8AZ48 A0A0P5QID4 A0A0P5WQL9 A0A336MNG2 A0A0P5BL96 A0A0P6E5F6 A0A0P5PI39 A0A0P5U4M3 A0A0P5SDM2 A0A0P5JRM0 A0A0P5XJS3 U5EVI2 A0A0P5WI39 A0A0P5K412 A0A0P5Q316 A0A1I8PI49 A0A0P5HVX7 A0A0P6CIX5 A0A0P5UWB3 A0A0N8BHX1 A0A2S2PBV2 A0A336L6N5 A0A0P5N2F5 A0A0P6GQH3 A0A0P5DE29 A0A0P5MWQ1 A0A0P5VQV1 A0A2H8TNL6 A0A195CPU1 A0A0P5UL62 A0A158NUX1 J9JMY8 A0A0P5S3D5 A0A0P5N233 A0A0P5SED1 W8B5Q2 A0A2S2QN44 A0A0P5EBY2 A0A0A1WM48 A0A0L0CB15 A0A0P6E034 A0A0P5MRB7 A0A0P6ENK1 A0A0P5S3D1 A0A0A1XIX7 A0A0P6AZ93 A0A232EN93 A0A0P6JTB9 A0A1Q3FLP0 A0A1Q3FLJ8 A0A1Q3FLB2 A0A1Q3FLM6 A0A1Q3FLM1 A0A1Q3FLL8 A0A023EWP9 E9IJA6 A0A0P5P545 B0W7R8 E2B0Y4 A0A1J1IC26 A0A1Q3FLG3 A0A1Q3FLU3 Q17KN9 A0A1Q3FLL7 A0A1Q3FLN6
A0A2A4JVZ9 A0A194QU26 Q86G88 A0A1E1WLE5 A0A0L7LQM8 A0A067QUQ4 A0A1L8E400 A0A1L8E3C4 A0A1L8E3J4 A0A026VUY3 A0A1B0GKX3 A0A2A3ECL3 A0A3L8DE97 F4X5Q1 A0A195FQK3 A0A088ACX9 A0A154P056 A0A310STA2 A0A0P5N3H1 A0A0N8BB58 A0A0P6GVW7 A0A0P4ZC31 E9HAH4 A0A0P5C057 A0A0P4XX18 A0A0N8CM66 A0A0N8BWE9 A0A195BWJ3 A0A0N8AZ48 A0A0P5QID4 A0A0P5WQL9 A0A336MNG2 A0A0P5BL96 A0A0P6E5F6 A0A0P5PI39 A0A0P5U4M3 A0A0P5SDM2 A0A0P5JRM0 A0A0P5XJS3 U5EVI2 A0A0P5WI39 A0A0P5K412 A0A0P5Q316 A0A1I8PI49 A0A0P5HVX7 A0A0P6CIX5 A0A0P5UWB3 A0A0N8BHX1 A0A2S2PBV2 A0A336L6N5 A0A0P5N2F5 A0A0P6GQH3 A0A0P5DE29 A0A0P5MWQ1 A0A0P5VQV1 A0A2H8TNL6 A0A195CPU1 A0A0P5UL62 A0A158NUX1 J9JMY8 A0A0P5S3D5 A0A0P5N233 A0A0P5SED1 W8B5Q2 A0A2S2QN44 A0A0P5EBY2 A0A0A1WM48 A0A0L0CB15 A0A0P6E034 A0A0P5MRB7 A0A0P6ENK1 A0A0P5S3D1 A0A0A1XIX7 A0A0P6AZ93 A0A232EN93 A0A0P6JTB9 A0A1Q3FLP0 A0A1Q3FLJ8 A0A1Q3FLB2 A0A1Q3FLM6 A0A1Q3FLM1 A0A1Q3FLL8 A0A023EWP9 E9IJA6 A0A0P5P545 B0W7R8 E2B0Y4 A0A1J1IC26 A0A1Q3FLG3 A0A1Q3FLU3 Q17KN9 A0A1Q3FLL7 A0A1Q3FLN6
Pubmed
EMBL
KJ511849
AII79408.1
BABH01013908
AGBW02012896
OWR44261.1
KQ459599
+ More
KPI94369.1 ODYU01001858 SOQ38646.1 GDQN01001494 JAT89560.1 NWSH01000571 PCG75570.1 KQ461154 KPJ08495.1 AY237585 AAO85803.1 GDQN01003259 JAT87795.1 JTDY01000323 KOB77704.1 KK853387 KDR07962.1 GFDF01000819 JAV13265.1 GFDF01000928 JAV13156.1 GFDF01000781 JAV13303.1 KK108839 EZA46609.1 AJWK01033902 KZ288285 PBC29468.1 QOIP01000009 RLU18616.1 GL888739 EGI58217.1 KQ981335 KYN42577.1 KQ434787 KZC05299.1 KQ760551 OAD59829.1 GDIQ01155257 JAK96468.1 GDIQ01194995 JAK56730.1 GDIQ01038349 JAN56388.1 GDIP01214610 JAJ08792.1 GL732612 EFX71180.1 GDIP01181316 GDIP01146221 GDIQ01162407 GDIP01069736 GDIQ01029605 JAJ42086.1 JAK89318.1 GDIP01236830 JAI86571.1 GDIP01118119 JAL85595.1 GDIQ01138267 JAL13459.1 KQ976401 KYM92650.1 GDIQ01228675 JAK23050.1 GDIQ01120213 JAL31513.1 GDIP01082984 JAM20731.1 UFQS01001739 UFQT01001739 SSX12129.1 SSX31590.1 GDIP01197920 JAJ25482.1 GDIP01146223 GDIP01050735 GDIQ01067840 JAJ77179.1 JAN26897.1 GDIQ01138266 JAL13460.1 GDIP01118121 JAL85593.1 GDIQ01095026 JAL56700.1 GDIQ01194994 JAK56731.1 GDIP01071331 JAM32384.1 GANO01001848 JAB58023.1 GDIP01086133 JAM17582.1 GDIQ01194996 JAK56729.1 GDIQ01120214 JAL31512.1 GDIQ01228074 JAK23651.1 GDIQ01090474 JAN04263.1 GDIP01107386 JAL96328.1 GDIQ01176091 JAK75634.1 GGMR01014243 MBY26862.1 SSX12128.1 SSX31589.1 GDIQ01147972 JAL03754.1 GDIQ01031151 JAN63586.1 GDIP01157716 JAJ65686.1 GDIQ01152408 JAK99317.1 GDIP01096788 JAM06927.1 GFXV01003930 MBW15735.1 KQ977444 KYN02758.1 GDIP01113680 JAL90034.1 ADTU01026806 ADTU01026807 ADTU01026808 ABLF02022377 ABLF02022379 GDIQ01092928 JAL58798.1 GDIQ01152407 JAK99318.1 GDIQ01092930 JAL58796.1 GAMC01021441 JAB85114.1 GGMS01009767 MBY78970.1 GDIP01162510 JAJ60892.1 GBXI01014168 GBXI01011409 GBXI01000867 JAD00124.1 JAD02883.1 JAD13425.1 JRES01000755 KNC28664.1 GDIQ01070438 JAN24299.1 GDIQ01152406 JAK99319.1 GDIQ01073182 JAN21555.1 GDIQ01092929 JAL58797.1 GBXI01003457 JAD10835.1 GDIP01022314 JAM81401.1 NNAY01003194 OXU19825.1 GDIQ01009218 JAN85519.1 GFDL01006590 JAV28455.1 GFDL01006651 JAV28394.1 GFDL01006645 JAV28400.1 GFDL01006621 JAV28424.1 GFDL01006610 JAV28435.1 GFDL01006637 JAV28408.1 GAPW01000065 JAC13533.1 GL763764 EFZ19346.1 GDIQ01133477 JAL18249.1 DS231855 EDS38212.1 GL444716 EFN60653.1 CVRI01000044 CRK96534.1 GFDL01006624 JAV28421.1 GFDL01006557 JAV28488.1 CH477222 EAT47265.1 GFDL01006623 JAV28422.1 GFDL01006554 JAV28491.1
KPI94369.1 ODYU01001858 SOQ38646.1 GDQN01001494 JAT89560.1 NWSH01000571 PCG75570.1 KQ461154 KPJ08495.1 AY237585 AAO85803.1 GDQN01003259 JAT87795.1 JTDY01000323 KOB77704.1 KK853387 KDR07962.1 GFDF01000819 JAV13265.1 GFDF01000928 JAV13156.1 GFDF01000781 JAV13303.1 KK108839 EZA46609.1 AJWK01033902 KZ288285 PBC29468.1 QOIP01000009 RLU18616.1 GL888739 EGI58217.1 KQ981335 KYN42577.1 KQ434787 KZC05299.1 KQ760551 OAD59829.1 GDIQ01155257 JAK96468.1 GDIQ01194995 JAK56730.1 GDIQ01038349 JAN56388.1 GDIP01214610 JAJ08792.1 GL732612 EFX71180.1 GDIP01181316 GDIP01146221 GDIQ01162407 GDIP01069736 GDIQ01029605 JAJ42086.1 JAK89318.1 GDIP01236830 JAI86571.1 GDIP01118119 JAL85595.1 GDIQ01138267 JAL13459.1 KQ976401 KYM92650.1 GDIQ01228675 JAK23050.1 GDIQ01120213 JAL31513.1 GDIP01082984 JAM20731.1 UFQS01001739 UFQT01001739 SSX12129.1 SSX31590.1 GDIP01197920 JAJ25482.1 GDIP01146223 GDIP01050735 GDIQ01067840 JAJ77179.1 JAN26897.1 GDIQ01138266 JAL13460.1 GDIP01118121 JAL85593.1 GDIQ01095026 JAL56700.1 GDIQ01194994 JAK56731.1 GDIP01071331 JAM32384.1 GANO01001848 JAB58023.1 GDIP01086133 JAM17582.1 GDIQ01194996 JAK56729.1 GDIQ01120214 JAL31512.1 GDIQ01228074 JAK23651.1 GDIQ01090474 JAN04263.1 GDIP01107386 JAL96328.1 GDIQ01176091 JAK75634.1 GGMR01014243 MBY26862.1 SSX12128.1 SSX31589.1 GDIQ01147972 JAL03754.1 GDIQ01031151 JAN63586.1 GDIP01157716 JAJ65686.1 GDIQ01152408 JAK99317.1 GDIP01096788 JAM06927.1 GFXV01003930 MBW15735.1 KQ977444 KYN02758.1 GDIP01113680 JAL90034.1 ADTU01026806 ADTU01026807 ADTU01026808 ABLF02022377 ABLF02022379 GDIQ01092928 JAL58798.1 GDIQ01152407 JAK99318.1 GDIQ01092930 JAL58796.1 GAMC01021441 JAB85114.1 GGMS01009767 MBY78970.1 GDIP01162510 JAJ60892.1 GBXI01014168 GBXI01011409 GBXI01000867 JAD00124.1 JAD02883.1 JAD13425.1 JRES01000755 KNC28664.1 GDIQ01070438 JAN24299.1 GDIQ01152406 JAK99319.1 GDIQ01073182 JAN21555.1 GDIQ01092929 JAL58797.1 GBXI01003457 JAD10835.1 GDIP01022314 JAM81401.1 NNAY01003194 OXU19825.1 GDIQ01009218 JAN85519.1 GFDL01006590 JAV28455.1 GFDL01006651 JAV28394.1 GFDL01006645 JAV28400.1 GFDL01006621 JAV28424.1 GFDL01006610 JAV28435.1 GFDL01006637 JAV28408.1 GAPW01000065 JAC13533.1 GL763764 EFZ19346.1 GDIQ01133477 JAL18249.1 DS231855 EDS38212.1 GL444716 EFN60653.1 CVRI01000044 CRK96534.1 GFDL01006624 JAV28421.1 GFDL01006557 JAV28488.1 CH477222 EAT47265.1 GFDL01006623 JAV28422.1 GFDL01006554 JAV28491.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000218220
UP000053240
UP000037510
+ More
UP000027135 UP000053097 UP000092461 UP000242457 UP000279307 UP000007755 UP000078541 UP000005203 UP000076502 UP000000305 UP000078540 UP000095300 UP000078542 UP000005205 UP000007819 UP000037069 UP000215335 UP000002320 UP000000311 UP000183832 UP000008820
UP000027135 UP000053097 UP000092461 UP000242457 UP000279307 UP000007755 UP000078541 UP000005203 UP000076502 UP000000305 UP000078540 UP000095300 UP000078542 UP000005205 UP000007819 UP000037069 UP000215335 UP000002320 UP000000311 UP000183832 UP000008820
PRIDE
Interpro
SUPFAM
SSF69179
SSF69179
Gene 3D
ProteinModelPortal
A0A076JVB3
H9J7I5
A0A212ES31
A0A194PNQ1
A0A2H1VCY1
A0A1E1WRC4
+ More
A0A2A4JVZ9 A0A194QU26 Q86G88 A0A1E1WLE5 A0A0L7LQM8 A0A067QUQ4 A0A1L8E400 A0A1L8E3C4 A0A1L8E3J4 A0A026VUY3 A0A1B0GKX3 A0A2A3ECL3 A0A3L8DE97 F4X5Q1 A0A195FQK3 A0A088ACX9 A0A154P056 A0A310STA2 A0A0P5N3H1 A0A0N8BB58 A0A0P6GVW7 A0A0P4ZC31 E9HAH4 A0A0P5C057 A0A0P4XX18 A0A0N8CM66 A0A0N8BWE9 A0A195BWJ3 A0A0N8AZ48 A0A0P5QID4 A0A0P5WQL9 A0A336MNG2 A0A0P5BL96 A0A0P6E5F6 A0A0P5PI39 A0A0P5U4M3 A0A0P5SDM2 A0A0P5JRM0 A0A0P5XJS3 U5EVI2 A0A0P5WI39 A0A0P5K412 A0A0P5Q316 A0A1I8PI49 A0A0P5HVX7 A0A0P6CIX5 A0A0P5UWB3 A0A0N8BHX1 A0A2S2PBV2 A0A336L6N5 A0A0P5N2F5 A0A0P6GQH3 A0A0P5DE29 A0A0P5MWQ1 A0A0P5VQV1 A0A2H8TNL6 A0A195CPU1 A0A0P5UL62 A0A158NUX1 J9JMY8 A0A0P5S3D5 A0A0P5N233 A0A0P5SED1 W8B5Q2 A0A2S2QN44 A0A0P5EBY2 A0A0A1WM48 A0A0L0CB15 A0A0P6E034 A0A0P5MRB7 A0A0P6ENK1 A0A0P5S3D1 A0A0A1XIX7 A0A0P6AZ93 A0A232EN93 A0A0P6JTB9 A0A1Q3FLP0 A0A1Q3FLJ8 A0A1Q3FLB2 A0A1Q3FLM6 A0A1Q3FLM1 A0A1Q3FLL8 A0A023EWP9 E9IJA6 A0A0P5P545 B0W7R8 E2B0Y4 A0A1J1IC26 A0A1Q3FLG3 A0A1Q3FLU3 Q17KN9 A0A1Q3FLL7 A0A1Q3FLN6
A0A2A4JVZ9 A0A194QU26 Q86G88 A0A1E1WLE5 A0A0L7LQM8 A0A067QUQ4 A0A1L8E400 A0A1L8E3C4 A0A1L8E3J4 A0A026VUY3 A0A1B0GKX3 A0A2A3ECL3 A0A3L8DE97 F4X5Q1 A0A195FQK3 A0A088ACX9 A0A154P056 A0A310STA2 A0A0P5N3H1 A0A0N8BB58 A0A0P6GVW7 A0A0P4ZC31 E9HAH4 A0A0P5C057 A0A0P4XX18 A0A0N8CM66 A0A0N8BWE9 A0A195BWJ3 A0A0N8AZ48 A0A0P5QID4 A0A0P5WQL9 A0A336MNG2 A0A0P5BL96 A0A0P6E5F6 A0A0P5PI39 A0A0P5U4M3 A0A0P5SDM2 A0A0P5JRM0 A0A0P5XJS3 U5EVI2 A0A0P5WI39 A0A0P5K412 A0A0P5Q316 A0A1I8PI49 A0A0P5HVX7 A0A0P6CIX5 A0A0P5UWB3 A0A0N8BHX1 A0A2S2PBV2 A0A336L6N5 A0A0P5N2F5 A0A0P6GQH3 A0A0P5DE29 A0A0P5MWQ1 A0A0P5VQV1 A0A2H8TNL6 A0A195CPU1 A0A0P5UL62 A0A158NUX1 J9JMY8 A0A0P5S3D5 A0A0P5N233 A0A0P5SED1 W8B5Q2 A0A2S2QN44 A0A0P5EBY2 A0A0A1WM48 A0A0L0CB15 A0A0P6E034 A0A0P5MRB7 A0A0P6ENK1 A0A0P5S3D1 A0A0A1XIX7 A0A0P6AZ93 A0A232EN93 A0A0P6JTB9 A0A1Q3FLP0 A0A1Q3FLJ8 A0A1Q3FLB2 A0A1Q3FLM6 A0A1Q3FLM1 A0A1Q3FLL8 A0A023EWP9 E9IJA6 A0A0P5P545 B0W7R8 E2B0Y4 A0A1J1IC26 A0A1Q3FLG3 A0A1Q3FLU3 Q17KN9 A0A1Q3FLL7 A0A1Q3FLN6
PDB
6DJP
E-value=3.03888e-77,
Score=738
Ontologies
GO
Topology
Subcellular location
Membrane
SignalP
Position: 1 - 16,
Likelihood: 0.627674
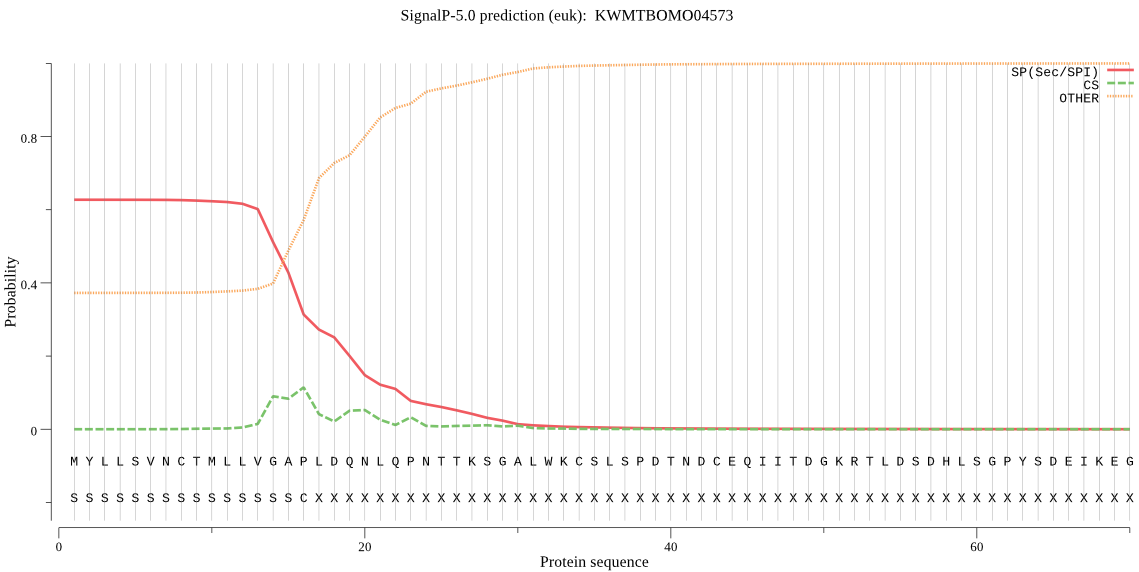
Length:
1063
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.91585
Exp number, first 60 AAs:
0.00345
Total prob of N-in:
0.00036
outside
1 - 1003
TMhelix
1004 - 1026
inside
1027 - 1063

Population Genetic Test Statistics
Pi
221.993945
Theta
173.259914
Tajima's D
0.897456
CLR
0.001338
CSRT
0.630868456577171
Interpretation
Uncertain
