Pre Gene Modal
BGIBMGA005473
Annotation
PREDICTED:_ATP-binding_cassette_transporter_subfamily_B_isoform_X1_[Bombyx_mori]
Full name
Guanylate cyclase
Location in the cell
PlasmaMembrane Reliability : 4.769
Sequence
CDS
ATGGTAGTTGCATTAACAGTTACTCTGGTAGTATTTCCATTGTCGGCCTACTTAGCGATCCTCGAGAGGAGGTTCTTATTACCGTCCGTGCCTCCTAGAGGTCACGGTTTTGTACTCTTGGTTTTTTGGGCTATGATATTCGTATCGGAGAATTTATCATTTTTAAATATCAACAAAGAAGGATGGTGGTGGCACTTGAAAAATCTTCAAGATCGCCTCGAAATGTCATTGTTCGTCGGTCGTTACGTGTCGTGTATGATAATGTTCGTGCTCGGTATGAAGGCGCCGGGCATCATGCACCACTTCGACTACTTAGAGGACGAACACGAAGGCCGAAGGAATATACAGCCACGCGATGACAACAGATCGACGTTCAGAAATGTGTTTACGAAATTGCGCACACTCCTACCGTTCCTGTGGCCGAAGAAGAATTTCATCCTCCAACTCTATGTGCTCATATGTATACTGGCTCTCATTGCTGGAAGAGCTATCAATCTATACGTCCCTATATACAACAGGCTGATATTTAACAGCATATCGGAACCGCCTCTCTCCTTCCGCTGGGATCTGGTATTGATATATGTGTTTTTCAAGTTCCTCCAAGGAGGGGGTACCGGCGGAATGGGTTTATTGAATAATATGCGTTCGTTTTTGTGGATCAAGGTCCAACAATACACCACAAGAGAGTTGGAGCTGGAACTGTTCAAGCATCTCCATGACTTGCCTTTACGCTGGCATTTGTCTCGCAAAACCGGAGAAGTCCTCAGGGTGATGGACCGAGGAACGGATTCTATAAACAATCTCCTCTCGTACATTCTGTTCTCTATCACTCCAACGCTGGTCGATATCTTGGTGGCAGTCATCTATTTTGTCACGGCGTTCAATATTTGGTTCGGACTGATCGTTTTTACTACAATGATCCTGTATATTATCGCAACAATAGCCGTGACGGAATGGCGCACGAAGTTCCAACGTCGCATGAATCTCGCCGACAACGAGCAGAAGGCTCGCTCGGTCGACTCTCTGTTGAACTACGAGACTGTTAAATATTACGGAGCGGAATCTTATGAAGTGATGTCTTATAGAGAAGCCATCCTTAATTATCAGAAAGAAGAGTTTAAATCATTGTTCACATTAAACATTTTGAACACCATTCAAAATATCATCATCTGCAGCGGCTTACTTGCTGGTTCGTTGCTCTGCGTGTCGTTGGTGGTGAAGACGGCTGAATTCACTGTCGGTGACTACGCTTTGTTCGCGTCGTACATAGTGCAGCTTTACGTTCCCTTGAATTGGTTCGGAACATATTACAGAGCTATACAGAAGAATTTCGTGGACATGGAGAACATGTTCGACCTGCTGAGGGTGGATACGGACGTGCGCGACGCGGCCGGGGCGCCCGAGCTGGTGGTGCGGCGCGGCGGAGTCGAGTTCAAGCACGTGTCCTTCGGATACGGCTCCGAGAGGCTCGTCCTTAACAACATCAGTTTTAAAATCGCCCCCGGAAGCACCGTTGCATTGGTCGGGCCCAGCGGCGCGGGAAAATCGACGATAATGCGTCTGTTATTCCGTTTTTACGACGTGAACGAGGGCGCCGTGCTGGTGGACGAGCAGGACGTGCGCACCGTCACGCAGGCCTCGCTGCGCGCCAACATCGGCGTCGTGCCGCAGGACACGGTGCTCTTCAACAACACCGTTAGATACAACATTCAATACGGAAGATTGAACGCGCCCGCCGCTGATATAATATCTGCCGCCAAGAACGCGGACATTCACGACCGAATACTGACGTTTCCCGACGCTTACGACACGCAGGTTGGAGAAAGGGGCCTTAGGCTGAGCGGTGGCGAAAAGCAAAGAATAGCTATTGCACGAACGATCTTAAAAGATCCCGCCATTGTCCTGCTCGATGAAGCTACTTCTGCGCTAGACACGAACACAGAAAGAAATATTCAAGCCGCGTTGGCACGAGTGTGCGCAAACAGAACTACCCTTATAATTGCACACAGGCTCTCAACGATCATTCATGCCGACGAAATACTGGTCCTTAAGGACGGTGAGATTATCGAACGTGGAAACCATGAAGCCCTCCTAGCGCAGGCCGGGTTCTATGCGTCCATGTGGCAACAGCAACTCGAGAATCGAAACAGCAACGGTAACAACGATGATGACGTCAGCGGCGGCGAAGGGAACAACAACCAGAGGAGCCAAATCGTCGCGCCCGTGTTCGGTCACGGCCATGGTCATGGGCATGGTCATTAA
Protein
MVVALTVTLVVFPLSAYLAILERRFLLPSVPPRGHGFVLLVFWAMIFVSENLSFLNINKEGWWWHLKNLQDRLEMSLFVGRYVSCMIMFVLGMKAPGIMHHFDYLEDEHEGRRNIQPRDDNRSTFRNVFTKLRTLLPFLWPKKNFILQLYVLICILALIAGRAINLYVPIYNRLIFNSISEPPLSFRWDLVLIYVFFKFLQGGGTGGMGLLNNMRSFLWIKVQQYTTRELELELFKHLHDLPLRWHLSRKTGEVLRVMDRGTDSINNLLSYILFSITPTLVDILVAVIYFVTAFNIWFGLIVFTTMILYIIATIAVTEWRTKFQRRMNLADNEQKARSVDSLLNYETVKYYGAESYEVMSYREAILNYQKEEFKSLFTLNILNTIQNIIICSGLLAGSLLCVSLVVKTAEFTVGDYALFASYIVQLYVPLNWFGTYYRAIQKNFVDMENMFDLLRVDTDVRDAAGAPELVVRRGGVEFKHVSFGYGSERLVLNNISFKIAPGSTVALVGPSGAGKSTIMRLLFRFYDVNEGAVLVDEQDVRTVTQASLRANIGVVPQDTVLFNNTVRYNIQYGRLNAPAADIISAAKNADIHDRILTFPDAYDTQVGERGLRLSGGEKQRIAIARTILKDPAIVLLDEATSALDTNTERNIQAALARVCANRTTLIIAHRLSTIIHADEILVLKDGEIIERGNHEALLAQAGFYASMWQQQLENRNSNGNNDDDVSGGEGNNNQRSQIVAPVFGHGHGHGHGH
Summary
Catalytic Activity
GTP = 3',5'-cyclic GMP + diphosphate
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Feature
chain Guanylate cyclase
Uniprot
H9J7I1
F2YDQ5
A0A2H1VR01
A0A194QU21
A0A194PN02
A0A2J7RS07
+ More
A0A2J7RS02 A0A1Y1K9V1 A0A182J8S9 A0A067QIY2 A0A182XLW2 A0A182LQD1 Q7QKD8 A0A182HS10 A0A088ARD0 A0A182UIB4 A0A182UDT7 A0A182UYY8 A0A1L8DUM0 A0A1L8DUM7 A0A182PEG5 A0A182MX05 V5GJI0 A0A0H5BVD5 K7IWV7 A0A2A3EBS9 A0A232FCS4 A0A182R7N0 Q16ZP7 A0A1S4FIQ7 A0A0U9HU18 A0A182NAA1 A0A2M3Z3S9 A0A2M3Z3H4 A0A182FBM7 A0A2M4AA77 A0A2M4BDR4 A0A182GIV6 A0A2M4BDQ9 A0A2M4BDS1 A0A2M4A9K2 A0A182QA77 A0A1Q3FIS0 E2AI03 A0A1Q3FJ49 A0A1Q3FI81 A0A1Q3FID3 D2A5D4 W5JI45 A0A026VXZ9 N6TZJ9 A0A1J1ITU1 A0A195DFL3 A0A158P2U1 A0A336MLG8 U5EX21 A0A336KJI7 U4UN61 A0A154PIV9 A0A0N0BG58 E2BRY8 A0A1I8QBP0 A0A0C9QIE0 T1EB34 B0XFW9 A0A0P4VKT9 A0A0L0CC45 A0A069DWZ9 A0A224XIP0 A0A0A1WT89 A0A3B0JPZ0 A0A023EUV0 A0A1A9YK51 A0A023F3I3 B4GEV0 B4PRJ6 A0A0A9YUP4 Q9VF20 T1I161 B4HLH1 A0A1A9WJI9 Q296Q1 B3P3W5 A0A146KT46 B3M387 A0A0V0G5P0 W8C2W4 B4LX86 T1PGC8 A0A1I8M0V8 A0A1A9UTM3 B4JT46 A0A1W4VB65 B4NBA7 A0A182W2J8 A0A034VPM6 A0A0K8TYR2 A0A0K8VRN2 A0A0M4EHK7 A0A3L8D578
A0A2J7RS02 A0A1Y1K9V1 A0A182J8S9 A0A067QIY2 A0A182XLW2 A0A182LQD1 Q7QKD8 A0A182HS10 A0A088ARD0 A0A182UIB4 A0A182UDT7 A0A182UYY8 A0A1L8DUM0 A0A1L8DUM7 A0A182PEG5 A0A182MX05 V5GJI0 A0A0H5BVD5 K7IWV7 A0A2A3EBS9 A0A232FCS4 A0A182R7N0 Q16ZP7 A0A1S4FIQ7 A0A0U9HU18 A0A182NAA1 A0A2M3Z3S9 A0A2M3Z3H4 A0A182FBM7 A0A2M4AA77 A0A2M4BDR4 A0A182GIV6 A0A2M4BDQ9 A0A2M4BDS1 A0A2M4A9K2 A0A182QA77 A0A1Q3FIS0 E2AI03 A0A1Q3FJ49 A0A1Q3FI81 A0A1Q3FID3 D2A5D4 W5JI45 A0A026VXZ9 N6TZJ9 A0A1J1ITU1 A0A195DFL3 A0A158P2U1 A0A336MLG8 U5EX21 A0A336KJI7 U4UN61 A0A154PIV9 A0A0N0BG58 E2BRY8 A0A1I8QBP0 A0A0C9QIE0 T1EB34 B0XFW9 A0A0P4VKT9 A0A0L0CC45 A0A069DWZ9 A0A224XIP0 A0A0A1WT89 A0A3B0JPZ0 A0A023EUV0 A0A1A9YK51 A0A023F3I3 B4GEV0 B4PRJ6 A0A0A9YUP4 Q9VF20 T1I161 B4HLH1 A0A1A9WJI9 Q296Q1 B3P3W5 A0A146KT46 B3M387 A0A0V0G5P0 W8C2W4 B4LX86 T1PGC8 A0A1I8M0V8 A0A1A9UTM3 B4JT46 A0A1W4VB65 B4NBA7 A0A182W2J8 A0A034VPM6 A0A0K8TYR2 A0A0K8VRN2 A0A0M4EHK7 A0A3L8D578
EC Number
4.6.1.2
Pubmed
19121390
26354079
28004739
24845553
20966253
12364791
+ More
14747013 17210077 25244985 20075255 28648823 17510324 26483478 20798317 18362917 19820115 20920257 23761445 24508170 23537049 21347285 27129103 26108605 26334808 25830018 24945155 25474469 17994087 17550304 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19001374 26109357 26109356 15632085 18057021 24495485 25315136 25348373 29121518 30249741
14747013 17210077 25244985 20075255 28648823 17510324 26483478 20798317 18362917 19820115 20920257 23761445 24508170 23537049 21347285 27129103 26108605 26334808 25830018 24945155 25474469 17994087 17550304 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19001374 26109357 26109356 15632085 18057021 24495485 25315136 25348373 29121518 30249741
EMBL
BABH01013894
BABH01013895
HQ896031
ADZ56943.1
ODYU01003917
SOQ43271.1
+ More
KQ461154 KPJ08490.1 KQ459599 KPI94373.1 NEVH01000596 PNF43620.1 PNF43621.1 GEZM01093597 JAV56226.1 KK853292 KDR08849.1 AAAB01008799 EAA03770.5 APCN01000232 GFDF01003957 JAV10127.1 GFDF01003956 JAV10128.1 AXCM01000557 GALX01004212 JAB64254.1 LK392616 CDS25232.1 AAZX01010983 KZ288308 PBC28742.1 NNAY01000459 OXU28249.1 CH477487 EAT40111.1 GARF01000036 JAC88909.1 GGFM01002404 MBW23155.1 GGFM01002316 MBW23067.1 GGFK01004376 MBW37697.1 GGFJ01002043 MBW51184.1 JXUM01066956 JXUM01066957 JXUM01066958 JXUM01066959 JXUM01066960 KQ562425 KXJ75945.1 GGFJ01002049 MBW51190.1 GGFJ01001970 MBW51111.1 GGFK01004165 MBW37486.1 AXCN02002617 GFDL01007545 JAV27500.1 GL439621 EFN66911.1 GFDL01007446 JAV27599.1 GFDL01007788 JAV27257.1 GFDL01007728 JAV27317.1 KQ971345 EFA05094.1 ADMH02001118 ETN64062.1 KK107597 EZA48612.1 APGK01045421 APGK01045422 APGK01045423 KB741036 ENN74715.1 CVRI01000057 CRL01945.1 KQ980903 KYN11632.1 ADTU01007480 UFQS01001404 UFQT01001404 SSX10719.1 SSX30401.1 GANO01001160 JAB58711.1 UFQS01000450 UFQT01000450 SSX04132.1 SSX24497.1 KB632394 ERL94567.1 KQ434931 KZC11789.1 KQ435791 KOX74318.1 GL450107 EFN81504.1 GBYB01003269 GBYB01006338 JAG73036.1 JAG76105.1 GAMD01000393 JAB01198.1 DS232967 EDS27088.1 GDKW01001630 JAI54965.1 JRES01000623 KNC29821.1 GBGD01000484 JAC88405.1 GFTR01008116 JAW08310.1 GBXI01012471 JAD01821.1 OUUW01000008 SPP84274.1 GAPW01000383 JAC13215.1 GBBI01002789 JAC15923.1 CH479182 EDW34135.1 CM000160 EDW97396.1 GBHO01006882 GBHO01006879 GBRD01014300 GDHC01007934 JAG36722.1 JAG36725.1 JAG51526.1 JAQ10695.1 AE014297 AY084209 EU571211 AAF55241.1 AAL89947.1 ACE60575.1 ACPB03016557 ACPB03016558 ACPB03016559 CH480815 EDW41991.1 CM000070 EAL28407.2 CH954181 EDV49071.1 GDHC01018968 JAP99660.1 CH902617 EDV43548.1 KPU80307.1 GECL01003410 JAP02714.1 GAMC01002992 JAC03564.1 CH940650 EDW66738.2 KA647744 AFP62373.1 CH916373 EDV94936.1 CH964232 EDW81071.1 KY849624 GAKP01014523 ATY74508.1 JAC44429.1 GDHF01032712 GDHF01026906 GDHF01005764 JAI19602.1 JAI25408.1 JAI46550.1 GDHF01010765 JAI41549.1 CP012526 ALC45884.1 QOIP01000013 RLU15386.1
KQ461154 KPJ08490.1 KQ459599 KPI94373.1 NEVH01000596 PNF43620.1 PNF43621.1 GEZM01093597 JAV56226.1 KK853292 KDR08849.1 AAAB01008799 EAA03770.5 APCN01000232 GFDF01003957 JAV10127.1 GFDF01003956 JAV10128.1 AXCM01000557 GALX01004212 JAB64254.1 LK392616 CDS25232.1 AAZX01010983 KZ288308 PBC28742.1 NNAY01000459 OXU28249.1 CH477487 EAT40111.1 GARF01000036 JAC88909.1 GGFM01002404 MBW23155.1 GGFM01002316 MBW23067.1 GGFK01004376 MBW37697.1 GGFJ01002043 MBW51184.1 JXUM01066956 JXUM01066957 JXUM01066958 JXUM01066959 JXUM01066960 KQ562425 KXJ75945.1 GGFJ01002049 MBW51190.1 GGFJ01001970 MBW51111.1 GGFK01004165 MBW37486.1 AXCN02002617 GFDL01007545 JAV27500.1 GL439621 EFN66911.1 GFDL01007446 JAV27599.1 GFDL01007788 JAV27257.1 GFDL01007728 JAV27317.1 KQ971345 EFA05094.1 ADMH02001118 ETN64062.1 KK107597 EZA48612.1 APGK01045421 APGK01045422 APGK01045423 KB741036 ENN74715.1 CVRI01000057 CRL01945.1 KQ980903 KYN11632.1 ADTU01007480 UFQS01001404 UFQT01001404 SSX10719.1 SSX30401.1 GANO01001160 JAB58711.1 UFQS01000450 UFQT01000450 SSX04132.1 SSX24497.1 KB632394 ERL94567.1 KQ434931 KZC11789.1 KQ435791 KOX74318.1 GL450107 EFN81504.1 GBYB01003269 GBYB01006338 JAG73036.1 JAG76105.1 GAMD01000393 JAB01198.1 DS232967 EDS27088.1 GDKW01001630 JAI54965.1 JRES01000623 KNC29821.1 GBGD01000484 JAC88405.1 GFTR01008116 JAW08310.1 GBXI01012471 JAD01821.1 OUUW01000008 SPP84274.1 GAPW01000383 JAC13215.1 GBBI01002789 JAC15923.1 CH479182 EDW34135.1 CM000160 EDW97396.1 GBHO01006882 GBHO01006879 GBRD01014300 GDHC01007934 JAG36722.1 JAG36725.1 JAG51526.1 JAQ10695.1 AE014297 AY084209 EU571211 AAF55241.1 AAL89947.1 ACE60575.1 ACPB03016557 ACPB03016558 ACPB03016559 CH480815 EDW41991.1 CM000070 EAL28407.2 CH954181 EDV49071.1 GDHC01018968 JAP99660.1 CH902617 EDV43548.1 KPU80307.1 GECL01003410 JAP02714.1 GAMC01002992 JAC03564.1 CH940650 EDW66738.2 KA647744 AFP62373.1 CH916373 EDV94936.1 CH964232 EDW81071.1 KY849624 GAKP01014523 ATY74508.1 JAC44429.1 GDHF01032712 GDHF01026906 GDHF01005764 JAI19602.1 JAI25408.1 JAI46550.1 GDHF01010765 JAI41549.1 CP012526 ALC45884.1 QOIP01000013 RLU15386.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000235965
UP000075880
UP000027135
+ More
UP000076407 UP000075882 UP000007062 UP000075840 UP000005203 UP000075902 UP000075903 UP000075885 UP000075883 UP000076408 UP000002358 UP000242457 UP000215335 UP000075900 UP000008820 UP000075884 UP000069272 UP000069940 UP000249989 UP000075886 UP000000311 UP000007266 UP000000673 UP000053097 UP000019118 UP000183832 UP000078492 UP000005205 UP000030742 UP000076502 UP000053105 UP000008237 UP000095300 UP000002320 UP000037069 UP000268350 UP000092443 UP000008744 UP000002282 UP000000803 UP000015103 UP000001292 UP000091820 UP000001819 UP000008711 UP000007801 UP000008792 UP000095301 UP000078200 UP000001070 UP000192221 UP000007798 UP000075920 UP000092553 UP000279307
UP000076407 UP000075882 UP000007062 UP000075840 UP000005203 UP000075902 UP000075903 UP000075885 UP000075883 UP000076408 UP000002358 UP000242457 UP000215335 UP000075900 UP000008820 UP000075884 UP000069272 UP000069940 UP000249989 UP000075886 UP000000311 UP000007266 UP000000673 UP000053097 UP000019118 UP000183832 UP000078492 UP000005205 UP000030742 UP000076502 UP000053105 UP000008237 UP000095300 UP000002320 UP000037069 UP000268350 UP000092443 UP000008744 UP000002282 UP000000803 UP000015103 UP000001292 UP000091820 UP000001819 UP000008711 UP000007801 UP000008792 UP000095301 UP000078200 UP000001070 UP000192221 UP000007798 UP000075920 UP000092553 UP000279307
Pfam
Interpro
IPR003593
AAA+_ATPase
+ More
IPR017871 ABC_transporter_CS
IPR039421 Type_I_exporter
IPR032410 MTABC_N
IPR003439 ABC_transporter-like
IPR036640 ABC1_TM_sf
IPR027417 P-loop_NTPase
IPR011527 ABC1_TM_dom
IPR036671 Znf_DHP_sf
IPR007872 Znf_DHP
IPR000719 Prot_kinase_dom
IPR029787 Nucleotide_cyclase
IPR001054 A/G_cyclase
IPR028082 Peripla_BP_I
IPR011009 Kinase-like_dom_sf
IPR001170 ANPR/GUC
IPR001828 ANF_lig-bd_rcpt
IPR018297 A/G_cyclase_CS
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR017871 ABC_transporter_CS
IPR039421 Type_I_exporter
IPR032410 MTABC_N
IPR003439 ABC_transporter-like
IPR036640 ABC1_TM_sf
IPR027417 P-loop_NTPase
IPR011527 ABC1_TM_dom
IPR036671 Znf_DHP_sf
IPR007872 Znf_DHP
IPR000719 Prot_kinase_dom
IPR029787 Nucleotide_cyclase
IPR001054 A/G_cyclase
IPR028082 Peripla_BP_I
IPR011009 Kinase-like_dom_sf
IPR001170 ANPR/GUC
IPR001828 ANF_lig-bd_rcpt
IPR018297 A/G_cyclase_CS
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
SUPFAM
Gene 3D
ProteinModelPortal
H9J7I1
F2YDQ5
A0A2H1VR01
A0A194QU21
A0A194PN02
A0A2J7RS07
+ More
A0A2J7RS02 A0A1Y1K9V1 A0A182J8S9 A0A067QIY2 A0A182XLW2 A0A182LQD1 Q7QKD8 A0A182HS10 A0A088ARD0 A0A182UIB4 A0A182UDT7 A0A182UYY8 A0A1L8DUM0 A0A1L8DUM7 A0A182PEG5 A0A182MX05 V5GJI0 A0A0H5BVD5 K7IWV7 A0A2A3EBS9 A0A232FCS4 A0A182R7N0 Q16ZP7 A0A1S4FIQ7 A0A0U9HU18 A0A182NAA1 A0A2M3Z3S9 A0A2M3Z3H4 A0A182FBM7 A0A2M4AA77 A0A2M4BDR4 A0A182GIV6 A0A2M4BDQ9 A0A2M4BDS1 A0A2M4A9K2 A0A182QA77 A0A1Q3FIS0 E2AI03 A0A1Q3FJ49 A0A1Q3FI81 A0A1Q3FID3 D2A5D4 W5JI45 A0A026VXZ9 N6TZJ9 A0A1J1ITU1 A0A195DFL3 A0A158P2U1 A0A336MLG8 U5EX21 A0A336KJI7 U4UN61 A0A154PIV9 A0A0N0BG58 E2BRY8 A0A1I8QBP0 A0A0C9QIE0 T1EB34 B0XFW9 A0A0P4VKT9 A0A0L0CC45 A0A069DWZ9 A0A224XIP0 A0A0A1WT89 A0A3B0JPZ0 A0A023EUV0 A0A1A9YK51 A0A023F3I3 B4GEV0 B4PRJ6 A0A0A9YUP4 Q9VF20 T1I161 B4HLH1 A0A1A9WJI9 Q296Q1 B3P3W5 A0A146KT46 B3M387 A0A0V0G5P0 W8C2W4 B4LX86 T1PGC8 A0A1I8M0V8 A0A1A9UTM3 B4JT46 A0A1W4VB65 B4NBA7 A0A182W2J8 A0A034VPM6 A0A0K8TYR2 A0A0K8VRN2 A0A0M4EHK7 A0A3L8D578
A0A2J7RS02 A0A1Y1K9V1 A0A182J8S9 A0A067QIY2 A0A182XLW2 A0A182LQD1 Q7QKD8 A0A182HS10 A0A088ARD0 A0A182UIB4 A0A182UDT7 A0A182UYY8 A0A1L8DUM0 A0A1L8DUM7 A0A182PEG5 A0A182MX05 V5GJI0 A0A0H5BVD5 K7IWV7 A0A2A3EBS9 A0A232FCS4 A0A182R7N0 Q16ZP7 A0A1S4FIQ7 A0A0U9HU18 A0A182NAA1 A0A2M3Z3S9 A0A2M3Z3H4 A0A182FBM7 A0A2M4AA77 A0A2M4BDR4 A0A182GIV6 A0A2M4BDQ9 A0A2M4BDS1 A0A2M4A9K2 A0A182QA77 A0A1Q3FIS0 E2AI03 A0A1Q3FJ49 A0A1Q3FI81 A0A1Q3FID3 D2A5D4 W5JI45 A0A026VXZ9 N6TZJ9 A0A1J1ITU1 A0A195DFL3 A0A158P2U1 A0A336MLG8 U5EX21 A0A336KJI7 U4UN61 A0A154PIV9 A0A0N0BG58 E2BRY8 A0A1I8QBP0 A0A0C9QIE0 T1EB34 B0XFW9 A0A0P4VKT9 A0A0L0CC45 A0A069DWZ9 A0A224XIP0 A0A0A1WT89 A0A3B0JPZ0 A0A023EUV0 A0A1A9YK51 A0A023F3I3 B4GEV0 B4PRJ6 A0A0A9YUP4 Q9VF20 T1I161 B4HLH1 A0A1A9WJI9 Q296Q1 B3P3W5 A0A146KT46 B3M387 A0A0V0G5P0 W8C2W4 B4LX86 T1PGC8 A0A1I8M0V8 A0A1A9UTM3 B4JT46 A0A1W4VB65 B4NBA7 A0A182W2J8 A0A034VPM6 A0A0K8TYR2 A0A0K8VRN2 A0A0M4EHK7 A0A3L8D578
PDB
4MRV
E-value=1.55558e-128,
Score=1179
Ontologies
GO
PANTHER
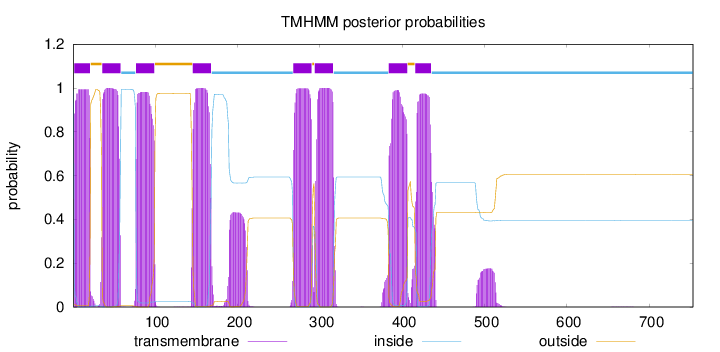
Topology
Length:
753
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
187.955370000001
Exp number, first 60 AAs:
41.73996
Total prob of N-in:
0.98826
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 21
outside
22 - 35
TMhelix
36 - 58
inside
59 - 76
TMhelix
77 - 99
outside
100 - 145
TMhelix
146 - 168
inside
169 - 267
TMhelix
268 - 290
outside
291 - 293
TMhelix
294 - 316
inside
317 - 383
TMhelix
384 - 406
outside
407 - 415
TMhelix
416 - 435
inside
436 - 753
Population Genetic Test Statistics
Pi
200.115252
Theta
155.162529
Tajima's D
1.095289
CLR
0.077233
CSRT
0.691715414229289
Interpretation
Uncertain
