Pre Gene Modal
BGIBMGA005335
Annotation
general_control_of_amino_acid_synthesis-like_protein_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.405 Nuclear Reliability : 1.1
Sequence
CDS
ATGCTGTCTTCTCTAATCAAAGAACATCAAGCAAAACAAGCAGCTAAACGTGTTGTACAAGAACAGAAACGTAAGGAGTGCATTACAGCTGCTAATGATTTAACACAAGCATTGGTAGATCATTTAAATGTCGGTGTTGCTCAAGCCTATTTAAATCAGAAAAAGTTAGATGCGGAAGCAAAACTTTTACACCAAGGTGCAATAAATTTTTCCAAACAAACACAGCAATGGTTGACTCTTGTGGAAAATTTCAGCAGTGCTCTCAAAGAAATTGGGGATGTCGAAAACTGGGCACGAAGTATAGAAAATGATATGAAAATTATTACAGACACTTTAGAAAGAGCATATGAAAAAGCACAAGAGAAACCAAGCAGTAGTCAATAA
Protein
MLSSLIKEHQAKQAAKRVVQEQKRKECITAANDLTQALVDHLNVGVAQAYLNQKKLDAEAKLLHQGAINFSKQTQQWLTLVENFSSALKEIGDVENWARSIENDMKIITDTLERAYEKAQEKPSSSQ
Summary
Uniprot
Q2F697
A0A1E1WDY5
A0A212EVE7
A0A3S2NEG7
A0A2A4IS66
A0A2H1V1B2
+ More
A0A194QY33 A0A0L7LAI2 S4PB37 H9J743 A0A1B6H852 A0A1B6G9U8 B0WME3 A0A067RH40 A0A0K8TS14 A0A2J7PPZ5 D6WZ78 A0A023EDX3 Q16ZK6 A0A1L8DGL2 A0A0P4VNT7 A0A224XPM4 A0A154PEX8 A0A1Q3FGY3 A0A084VRW6 A0A182MG19 A0A182UYM3 A0A182JQS6 A0A182UEI3 A0A182VQX5 A0A182XBM2 Q7PYD7 A0A182I0F6 A0A1I8MHS5 A0A0A9YMZ8 A0A023F5E4 A0A182FQK8 A0A182YJU7 A0A2H8TUJ5 A0A2S2NC93 C4WVJ5 A0A182PC19 A0A0C9R8X4 K7J0W5 A0A182J5L2 A0A1B6DY89 A0A1S3D6W6 T1DHM3 A0A2M4AXW3 A0A2M4C3T1 A0A2A3EEX2 V9IMR6 A0A088A9D9 A0A0L0CME9 W5J2B6 A0A1I8PQ02 A0A2S2QI89 A0A0L7R6L9 A0A1A9W7S2 A0A2R7W5L0 E2BEJ7 E0VK72 A0A1Y1MLI9 A0A026WWU6 A0A1Q3FGZ8 A0A1B0DPJ7 A0A1W4X1B6 A0A0C9RQ19 A0A158NFJ4 A0A1A9XBA4 A0A0K8U6U3 A0A0J7NS23 A0A0C9R635 W8AZ28 A0A0M4E5X6 U5EG57 B4KP11 E9ITG9 A0A1A9UWJ6 A0A1A9Z5G0 A0A151WNW7 A0A195D766 A0A0N0U3J2 B4LLN0 A0A1B0C080 A0A0T6AW07 A0A232FFR3 E9FYZ8 A0A0P6BN70 A0A1L8DG67 A0A164Y979 A0A0N8ANW6 A0A1B0CDF2 A0A151JYB8 F4X194 B3MEH5 A0A1W4UJJ0 A0A195DPF5 A0A2L2Y7I4 T1E3A4
A0A194QY33 A0A0L7LAI2 S4PB37 H9J743 A0A1B6H852 A0A1B6G9U8 B0WME3 A0A067RH40 A0A0K8TS14 A0A2J7PPZ5 D6WZ78 A0A023EDX3 Q16ZK6 A0A1L8DGL2 A0A0P4VNT7 A0A224XPM4 A0A154PEX8 A0A1Q3FGY3 A0A084VRW6 A0A182MG19 A0A182UYM3 A0A182JQS6 A0A182UEI3 A0A182VQX5 A0A182XBM2 Q7PYD7 A0A182I0F6 A0A1I8MHS5 A0A0A9YMZ8 A0A023F5E4 A0A182FQK8 A0A182YJU7 A0A2H8TUJ5 A0A2S2NC93 C4WVJ5 A0A182PC19 A0A0C9R8X4 K7J0W5 A0A182J5L2 A0A1B6DY89 A0A1S3D6W6 T1DHM3 A0A2M4AXW3 A0A2M4C3T1 A0A2A3EEX2 V9IMR6 A0A088A9D9 A0A0L0CME9 W5J2B6 A0A1I8PQ02 A0A2S2QI89 A0A0L7R6L9 A0A1A9W7S2 A0A2R7W5L0 E2BEJ7 E0VK72 A0A1Y1MLI9 A0A026WWU6 A0A1Q3FGZ8 A0A1B0DPJ7 A0A1W4X1B6 A0A0C9RQ19 A0A158NFJ4 A0A1A9XBA4 A0A0K8U6U3 A0A0J7NS23 A0A0C9R635 W8AZ28 A0A0M4E5X6 U5EG57 B4KP11 E9ITG9 A0A1A9UWJ6 A0A1A9Z5G0 A0A151WNW7 A0A195D766 A0A0N0U3J2 B4LLN0 A0A1B0C080 A0A0T6AW07 A0A232FFR3 E9FYZ8 A0A0P6BN70 A0A1L8DG67 A0A164Y979 A0A0N8ANW6 A0A1B0CDF2 A0A151JYB8 F4X194 B3MEH5 A0A1W4UJJ0 A0A195DPF5 A0A2L2Y7I4 T1E3A4
Pubmed
22118469
26354079
26227816
23622113
19121390
24845553
+ More
26369729 18362917 19820115 24945155 17510324 27129103 24438588 12364791 14747013 17210077 25315136 25401762 26823975 25474469 25244985 20075255 26108605 20920257 23761445 20798317 20566863 28004739 24508170 30249741 21347285 24495485 17994087 21282665 28648823 21292972 21719571 26561354 24330624
26369729 18362917 19820115 24945155 17510324 27129103 24438588 12364791 14747013 17210077 25315136 25401762 26823975 25474469 25244985 20075255 26108605 20920257 23761445 20798317 20566863 28004739 24508170 30249741 21347285 24495485 17994087 21282665 28648823 21292972 21719571 26561354 24330624
EMBL
DQ311175
ABD36120.1
GDQN01005844
JAT85210.1
AGBW02012201
OWR45465.1
+ More
RSAL01000163 RVE45356.1 NWSH01008373 PCG62581.1 ODYU01000221 SOQ34627.1 KQ461154 KPJ08486.1 JTDY01001936 KOB72503.1 GAIX01006132 JAA86428.1 BABH01013892 GECU01036828 JAS70878.1 GECZ01010569 GECZ01008022 GECZ01005894 JAS59200.1 JAS61747.1 JAS63875.1 DS231997 EDS31000.1 KK852510 KDR22358.1 GDAI01000464 JAI17139.1 NEVH01022641 PNF18405.1 KQ971372 EFA10392.1 GAPW01006096 JAC07502.1 CH477489 EAT40068.1 GFDF01008557 JAV05527.1 GDKW01002452 JAI54143.1 GFTR01001989 JAW14437.1 KQ434890 KZC10436.1 GFDL01008229 JAV26816.1 ATLV01015783 KE525036 KFB40710.1 AXCM01000371 AAAB01008987 EAA01678.4 APCN01002074 GBHO01009177 GBHO01009176 GBRD01013219 GDHC01006016 JAG34427.1 JAG34428.1 JAG52607.1 JAQ12613.1 GBBI01002359 JAC16353.1 GFXV01005437 MBW17242.1 GGMR01002146 MBY14765.1 ABLF02024690 AK341545 BAH71915.1 GBYB01009267 JAG79034.1 GEDC01006646 JAS30652.1 GAMD01002271 JAA99319.1 GGFK01012325 MBW45646.1 GGFJ01010768 MBW59909.1 KZ288277 PBC29716.1 JR053802 AEY62021.1 JRES01000195 KNC33441.1 ADMH02002134 ETN58392.1 GGMS01008210 MBY77413.1 KQ414646 KOC66537.1 KK854233 PTY13495.1 GL447817 EFN85887.1 DS235239 EEB13778.1 GEZM01032213 JAV84766.1 KK107079 QOIP01000010 EZA60186.1 RLU17442.1 GFDL01008218 JAV26827.1 AJVK01008120 AJVK01008121 GBYB01009266 JAG79033.1 ADTU01014529 GDHF01030016 JAI22298.1 LBMM01002123 KMQ95245.1 GBYB01002236 JAG72003.1 GAMC01012495 JAB94060.1 CP012524 ALC41778.1 GANO01003583 JAB56288.1 CH933808 EDW10077.1 GL765555 EFZ16108.1 KQ982893 KYQ49579.1 KQ976750 KYN08703.1 KQ435896 KOX69308.1 CH940648 EDW60893.1 JXJN01023495 LJIG01022686 KRT79256.1 NNAY01000332 OXU29147.1 GL732527 EFX87620.1 GDIP01013894 JAM89821.1 GFDF01008636 JAV05448.1 LRGB01000930 KZS15009.1 GDIQ01257331 JAJ94393.1 AJWK01007762 KQ981494 KYN40950.1 GL888528 EGI59766.1 CH902619 EDV37595.1 KQ980662 KYN14692.1 IAAA01014908 LAA04136.1 GALA01000019 JAA94833.1
RSAL01000163 RVE45356.1 NWSH01008373 PCG62581.1 ODYU01000221 SOQ34627.1 KQ461154 KPJ08486.1 JTDY01001936 KOB72503.1 GAIX01006132 JAA86428.1 BABH01013892 GECU01036828 JAS70878.1 GECZ01010569 GECZ01008022 GECZ01005894 JAS59200.1 JAS61747.1 JAS63875.1 DS231997 EDS31000.1 KK852510 KDR22358.1 GDAI01000464 JAI17139.1 NEVH01022641 PNF18405.1 KQ971372 EFA10392.1 GAPW01006096 JAC07502.1 CH477489 EAT40068.1 GFDF01008557 JAV05527.1 GDKW01002452 JAI54143.1 GFTR01001989 JAW14437.1 KQ434890 KZC10436.1 GFDL01008229 JAV26816.1 ATLV01015783 KE525036 KFB40710.1 AXCM01000371 AAAB01008987 EAA01678.4 APCN01002074 GBHO01009177 GBHO01009176 GBRD01013219 GDHC01006016 JAG34427.1 JAG34428.1 JAG52607.1 JAQ12613.1 GBBI01002359 JAC16353.1 GFXV01005437 MBW17242.1 GGMR01002146 MBY14765.1 ABLF02024690 AK341545 BAH71915.1 GBYB01009267 JAG79034.1 GEDC01006646 JAS30652.1 GAMD01002271 JAA99319.1 GGFK01012325 MBW45646.1 GGFJ01010768 MBW59909.1 KZ288277 PBC29716.1 JR053802 AEY62021.1 JRES01000195 KNC33441.1 ADMH02002134 ETN58392.1 GGMS01008210 MBY77413.1 KQ414646 KOC66537.1 KK854233 PTY13495.1 GL447817 EFN85887.1 DS235239 EEB13778.1 GEZM01032213 JAV84766.1 KK107079 QOIP01000010 EZA60186.1 RLU17442.1 GFDL01008218 JAV26827.1 AJVK01008120 AJVK01008121 GBYB01009266 JAG79033.1 ADTU01014529 GDHF01030016 JAI22298.1 LBMM01002123 KMQ95245.1 GBYB01002236 JAG72003.1 GAMC01012495 JAB94060.1 CP012524 ALC41778.1 GANO01003583 JAB56288.1 CH933808 EDW10077.1 GL765555 EFZ16108.1 KQ982893 KYQ49579.1 KQ976750 KYN08703.1 KQ435896 KOX69308.1 CH940648 EDW60893.1 JXJN01023495 LJIG01022686 KRT79256.1 NNAY01000332 OXU29147.1 GL732527 EFX87620.1 GDIP01013894 JAM89821.1 GFDF01008636 JAV05448.1 LRGB01000930 KZS15009.1 GDIQ01257331 JAJ94393.1 AJWK01007762 KQ981494 KYN40950.1 GL888528 EGI59766.1 CH902619 EDV37595.1 KQ980662 KYN14692.1 IAAA01014908 LAA04136.1 GALA01000019 JAA94833.1
Proteomes
UP000007151
UP000283053
UP000218220
UP000053240
UP000037510
UP000005204
+ More
UP000002320 UP000027135 UP000235965 UP000007266 UP000008820 UP000076502 UP000030765 UP000075883 UP000075903 UP000075881 UP000075902 UP000075920 UP000076407 UP000007062 UP000075840 UP000095301 UP000069272 UP000076408 UP000007819 UP000075885 UP000002358 UP000075880 UP000079169 UP000242457 UP000005203 UP000037069 UP000000673 UP000095300 UP000053825 UP000091820 UP000008237 UP000009046 UP000053097 UP000279307 UP000092462 UP000192223 UP000005205 UP000092443 UP000036403 UP000092553 UP000009192 UP000078200 UP000092445 UP000075809 UP000078542 UP000053105 UP000008792 UP000092460 UP000215335 UP000000305 UP000076858 UP000092461 UP000078541 UP000007755 UP000007801 UP000192221 UP000078492
UP000002320 UP000027135 UP000235965 UP000007266 UP000008820 UP000076502 UP000030765 UP000075883 UP000075903 UP000075881 UP000075902 UP000075920 UP000076407 UP000007062 UP000075840 UP000095301 UP000069272 UP000076408 UP000007819 UP000075885 UP000002358 UP000075880 UP000079169 UP000242457 UP000005203 UP000037069 UP000000673 UP000095300 UP000053825 UP000091820 UP000008237 UP000009046 UP000053097 UP000279307 UP000092462 UP000192223 UP000005205 UP000092443 UP000036403 UP000092553 UP000009192 UP000078200 UP000092445 UP000075809 UP000078542 UP000053105 UP000008792 UP000092460 UP000215335 UP000000305 UP000076858 UP000092461 UP000078541 UP000007755 UP000007801 UP000192221 UP000078492
Interpro
SUPFAM
SSF53613
SSF53613
Gene 3D
ProteinModelPortal
Q2F697
A0A1E1WDY5
A0A212EVE7
A0A3S2NEG7
A0A2A4IS66
A0A2H1V1B2
+ More
A0A194QY33 A0A0L7LAI2 S4PB37 H9J743 A0A1B6H852 A0A1B6G9U8 B0WME3 A0A067RH40 A0A0K8TS14 A0A2J7PPZ5 D6WZ78 A0A023EDX3 Q16ZK6 A0A1L8DGL2 A0A0P4VNT7 A0A224XPM4 A0A154PEX8 A0A1Q3FGY3 A0A084VRW6 A0A182MG19 A0A182UYM3 A0A182JQS6 A0A182UEI3 A0A182VQX5 A0A182XBM2 Q7PYD7 A0A182I0F6 A0A1I8MHS5 A0A0A9YMZ8 A0A023F5E4 A0A182FQK8 A0A182YJU7 A0A2H8TUJ5 A0A2S2NC93 C4WVJ5 A0A182PC19 A0A0C9R8X4 K7J0W5 A0A182J5L2 A0A1B6DY89 A0A1S3D6W6 T1DHM3 A0A2M4AXW3 A0A2M4C3T1 A0A2A3EEX2 V9IMR6 A0A088A9D9 A0A0L0CME9 W5J2B6 A0A1I8PQ02 A0A2S2QI89 A0A0L7R6L9 A0A1A9W7S2 A0A2R7W5L0 E2BEJ7 E0VK72 A0A1Y1MLI9 A0A026WWU6 A0A1Q3FGZ8 A0A1B0DPJ7 A0A1W4X1B6 A0A0C9RQ19 A0A158NFJ4 A0A1A9XBA4 A0A0K8U6U3 A0A0J7NS23 A0A0C9R635 W8AZ28 A0A0M4E5X6 U5EG57 B4KP11 E9ITG9 A0A1A9UWJ6 A0A1A9Z5G0 A0A151WNW7 A0A195D766 A0A0N0U3J2 B4LLN0 A0A1B0C080 A0A0T6AW07 A0A232FFR3 E9FYZ8 A0A0P6BN70 A0A1L8DG67 A0A164Y979 A0A0N8ANW6 A0A1B0CDF2 A0A151JYB8 F4X194 B3MEH5 A0A1W4UJJ0 A0A195DPF5 A0A2L2Y7I4 T1E3A4
A0A194QY33 A0A0L7LAI2 S4PB37 H9J743 A0A1B6H852 A0A1B6G9U8 B0WME3 A0A067RH40 A0A0K8TS14 A0A2J7PPZ5 D6WZ78 A0A023EDX3 Q16ZK6 A0A1L8DGL2 A0A0P4VNT7 A0A224XPM4 A0A154PEX8 A0A1Q3FGY3 A0A084VRW6 A0A182MG19 A0A182UYM3 A0A182JQS6 A0A182UEI3 A0A182VQX5 A0A182XBM2 Q7PYD7 A0A182I0F6 A0A1I8MHS5 A0A0A9YMZ8 A0A023F5E4 A0A182FQK8 A0A182YJU7 A0A2H8TUJ5 A0A2S2NC93 C4WVJ5 A0A182PC19 A0A0C9R8X4 K7J0W5 A0A182J5L2 A0A1B6DY89 A0A1S3D6W6 T1DHM3 A0A2M4AXW3 A0A2M4C3T1 A0A2A3EEX2 V9IMR6 A0A088A9D9 A0A0L0CME9 W5J2B6 A0A1I8PQ02 A0A2S2QI89 A0A0L7R6L9 A0A1A9W7S2 A0A2R7W5L0 E2BEJ7 E0VK72 A0A1Y1MLI9 A0A026WWU6 A0A1Q3FGZ8 A0A1B0DPJ7 A0A1W4X1B6 A0A0C9RQ19 A0A158NFJ4 A0A1A9XBA4 A0A0K8U6U3 A0A0J7NS23 A0A0C9R635 W8AZ28 A0A0M4E5X6 U5EG57 B4KP11 E9ITG9 A0A1A9UWJ6 A0A1A9Z5G0 A0A151WNW7 A0A195D766 A0A0N0U3J2 B4LLN0 A0A1B0C080 A0A0T6AW07 A0A232FFR3 E9FYZ8 A0A0P6BN70 A0A1L8DG67 A0A164Y979 A0A0N8ANW6 A0A1B0CDF2 A0A151JYB8 F4X194 B3MEH5 A0A1W4UJJ0 A0A195DPF5 A0A2L2Y7I4 T1E3A4
Ontologies
GO
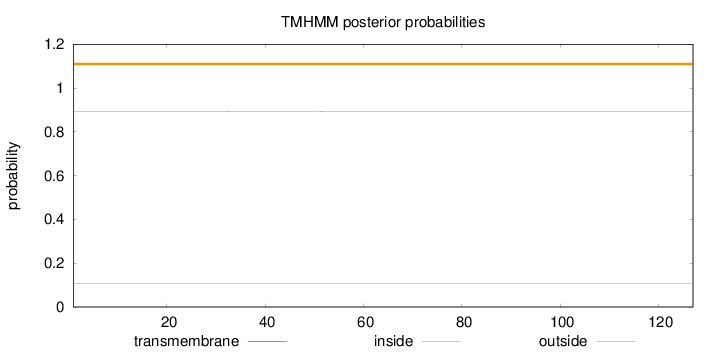
Topology
Length:
127
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00042
Exp number, first 60 AAs:
0.00042
Total prob of N-in:
0.10770
outside
1 - 127
Population Genetic Test Statistics
Pi
214.2673
Theta
143.764524
Tajima's D
1.131349
CLR
0.234559
CSRT
0.691465426728664
Interpretation
Uncertain
