Pre Gene Modal
BGIBMGA005337
Annotation
PREDICTED:_kazrin_isoform_X8_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.556
Sequence
CDS
ATGGCGCTAGTGCGCCGCATTCTAGGCGACGCCCAGGCGAAACTACGAAAAATGGTCGATGAGCAAGTCTCGGTGGGTACAAGGGTGGACGCTGACCCTGAACCAGAGCCCGCTGATCCTCTCATCACACCCGCTAGTTCTGTCCTGGACAGATCTGACTTGGACCACAGACCGATACCTCCTGAGCGCCGATTGCTGATAGGAACACTCGACAAACCTCGTAACGGATCTCTTAACGCTGACAAAGATGCCGGCACACTTAATAGAACCACACGATTAGCGATCAGAAATGGCAAAGATAATCCTTTTATAGACGATGACAACAAAGAAAACCAAGCCAATGGGAAACTTGATTCTCCAGTGAAATGGTCCACTCAAAACGGCAGCGATAGTGGAACATACGCTGAGATAGGTAATGGAAGTAACAGTAATGAAAGAAATATCCTGGACCCGGCCGATAGCTCTGAGGAAGAAGTTCCTCTGCCTGATGCTACCTCTCCTGGGAGAAGCAGCGATGGGCCAGAGCCGCGAGCTGATATTACAGCCACCCTTGAAGAGGCTGCATCACCAGGCGCTCGTTCGGACAGATCTCGTCAAGAGGAAATACCGGCATCCCCTGGGATGCTAACAGGGGCGCAATTAGCTCGCCGGCTTAGAATGGAAAACGAACGATTACAGGCGGAATTAACAAGAGTACGCCGACTACTGGTGGCAGCTGCTGAAAGGGGAGAAGCTGGCAACGCTCTTCTCGAGAGAACAAAAGATGATGAATCACAAGAAACAATCGCTTTGGACCCTCAGCAGCTTGAAAAGGAACTGCTCTTAGCTAGAGAAGCTGTCAATTCTCTAAAGGCGGATAAGAAAAGGTTAAAAGCAGAGAAATTCGATTTGTTGAATCAAATGAAACAACTCTACGCTACGCTTGAAGACAAAGAGAAGGAATTGCGTGATTTCATAAGGAATTATGAACAGATGCGTTCAAGAGGTGGCGCTTCTTCTGCTCTCGGAGCGGAAAGAGCAGAGAGGGAGCGTGAGCGGGCCGCACTCTTACGGCACGCTAGGGATGAGGCCGAGCGAAGTTTGCAACTGGCCGCTGCACTCAGCGCAAGGGACACTCAACTCAGACATGCCAGAGAACAACTCTTTGAGGCAAGACGACAGCTACAAGCAGCCGGGTGTCTGTCTGAAGGCGAAAGTGTGGCATCACTAGGCATCGGACCTCCGATGTTATTGGGAGGACCTACAGGTCTGATCGGGGACAGAGGCAGTTGTAGTGCAGATTCTGGAGTAAGAGGTAGCAGCGATGGAGGTGCCACTTCGGTATGCGGAGGCAACCTATCAGATTCCACAGCGGAAGGATTACCTCCGACTATTGACTCATATGACACCGATGGTGCGTCTTTGGTATCATCCGCGCATCACATTTATCAATTGGGCACTCCACGAGACTGCAGTCCTACAGTGTCACCGCACAACAGCGGATCGCCATTCACACGATCAATTGATGCAGGATCATTGTCAAGGTCAGTGGAACAGCTATCGAGCCCTGGTGAATGTGAGGCAGCACTCGTGGGGGGTCTGCGAGGACGCTCGGGGGGCAGCGCGGGAAGTAAAGGTGCCCGTGGTCGAGGTTCTGCGTGGGGATCCATATCCCGCGTATTTGCAAGGAGCCGACACCGTACCAAATCCGGCAGCGCTGCTGTGAGCGGTCATGAAAGTGAACCAGTATACGCGGGTACCGGTAGTACGACCCGTGCATGGTCGCCGCTAAGTAGTGAAGCAGCTCTCCGCGAGGCCGCTTCCCAGCCTTTAGCAAGGTGGCGCACACCGGCAATCATAGCGTGGCTAGAACTGGCTTTAGGAATGCCACAATATGCAGCTGCAATAGCCGATAACGTCAAAAGTGGCAAGGTTCTGCTCGAGTTGACGGATGCAGATCTGGAAGTCGGACTGGGGATCTCACAGCCGATGCACCGCAAGAAGCTGCGTCTTGCTATCGAAGAACGAAGACGACCAGACTTGGTCAGGAATCCAACAATAGGTCAACTCAGCCACGCTTGGGTCGCCGCGGAATGGTTACCAGATTTGGGCCTTTCTCAGTATGCTGAGTCATTCCTGGCTAACCTGGTGGATGCCCGCATGCTCGACACTATAAGCAAGAAAGAGTTAGAGAAATACCTCGGAGTTACAAGGAAGTTCCATCAGGCCTCCATTGTTCACGGCATTCATCTGCTGAGGATAATGAAATACGATCGACAAGCTCTGGCGGTGAGACGTCACCAGTGTGAGAACGTAGACGCGGACCCCCTGGTTTGGACCAATCACAGATTTATGCGGTGGGCCCATAATATTGATTTAGGTGAATTCGCTGACAATCTCAAAGATAGTGGAGTTCATGGCGGCCTGGTAGTACTGGAGCCGTCTTTCACCGGAGAAACAATGGCCACTGCTCTGGGTATTCCACCGTCTAAGAGCATTATTAGGAGACATTTAGTGGCTGAGTTTGATGCCCTTGTAATTCCTGCGAGAAACATGTTTGGCCACCAAATAAGAATTTTGGGGAGACCATTTTCAAGGTCTGTTGCCACTGGATTACCTGGGATTGACCTCAGCGGAGAGTCAAGGCGACATAGTCTACGGGGCTCAATAACTCGTGCTCTAGGGGTACTTAAGCCTAAACACGAACGAACATCCCCTTCAAGTTCTAGCGAGAGCTCCAGCGTGCTCAGTCTCGGTACTCAATCATATTCGTACTCACCTCCTGTGGCGGTACGAACCCTCTCTCAGCTGAGCATGCAGTACGCGCCGCCGCCGACCCTCCAGGAATACGAACCGATATACACACCATTGAGCCTGTATTCTCAATCCAGCGTCTCAACAAGGGACAGCCTACAAAGATTGAATGAAATCAAAGAGAATTCCAACATCATGCACAGGTACGGACAGAGAGCTGATCAAGCCCACAGAGTGAGTTCCCCTCTACCCAGTAAGTCCGTAGACGACTCCCTCGTCAAACAGAAGAGACACAGACGAGTCAAAAGCATTGGCGATATTAACGCTTCCAGTAAAACTTCTGTTTAG
Protein
MALVRRILGDAQAKLRKMVDEQVSVGTRVDADPEPEPADPLITPASSVLDRSDLDHRPIPPERRLLIGTLDKPRNGSLNADKDAGTLNRTTRLAIRNGKDNPFIDDDNKENQANGKLDSPVKWSTQNGSDSGTYAEIGNGSNSNERNILDPADSSEEEVPLPDATSPGRSSDGPEPRADITATLEEAASPGARSDRSRQEEIPASPGMLTGAQLARRLRMENERLQAELTRVRRLLVAAAERGEAGNALLERTKDDESQETIALDPQQLEKELLLAREAVNSLKADKKRLKAEKFDLLNQMKQLYATLEDKEKELRDFIRNYEQMRSRGGASSALGAERAERERERAALLRHARDEAERSLQLAAALSARDTQLRHAREQLFEARRQLQAAGCLSEGESVASLGIGPPMLLGGPTGLIGDRGSCSADSGVRGSSDGGATSVCGGNLSDSTAEGLPPTIDSYDTDGASLVSSAHHIYQLGTPRDCSPTVSPHNSGSPFTRSIDAGSLSRSVEQLSSPGECEAALVGGLRGRSGGSAGSKGARGRGSAWGSISRVFARSRHRTKSGSAAVSGHESEPVYAGTGSTTRAWSPLSSEAALREAASQPLARWRTPAIIAWLELALGMPQYAAAIADNVKSGKVLLELTDADLEVGLGISQPMHRKKLRLAIEERRRPDLVRNPTIGQLSHAWVAAEWLPDLGLSQYAESFLANLVDARMLDTISKKELEKYLGVTRKFHQASIVHGIHLLRIMKYDRQALAVRRHQCENVDADPLVWTNHRFMRWAHNIDLGEFADNLKDSGVHGGLVVLEPSFTGETMATALGIPPSKSIIRRHLVAEFDALVIPARNMFGHQIRILGRPFSRSVATGLPGIDLSGESRRHSLRGSITRALGVLKPKHERTSPSSSSESSSVLSLGTQSYSYSPPVAVRTLSQLSMQYAPPPTLQEYEPIYTPLSLYSQSSVSTRDSLQRLNEIKENSNIMHRYGQRADQAHRVSSPLPSKSVDDSLVKQKRHRRVKSIGDINASSKTSV
Summary
Uniprot
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF47769
SSF47769
Gene 3D
ProteinModelPortal
PDB
3TAD
E-value=9.95279e-49,
Score=492
Ontologies
GO
PANTHER
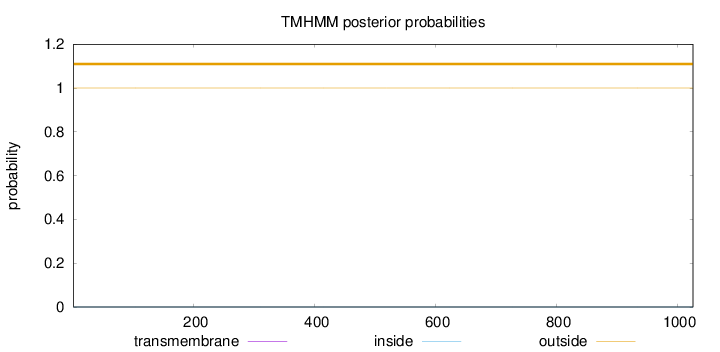
Topology
Length:
1026
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00159
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00001
outside
1 - 1026
Population Genetic Test Statistics
Pi
220.615171
Theta
180.391253
Tajima's D
0.780973
CLR
0.150749
CSRT
0.595670216489176
Interpretation
Uncertain
