Gene
KWMTBOMO04557
Annotation
PREDICTED:_uncharacterized_protein_LOC106141184_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.471 Nuclear Reliability : 2.029
Sequence
CDS
ATGAGAACTATAATTGCGTACTCTCAGGACATTCAAGAGGAGATGGATCGGCCCTCGCCCGTCAGATGCGACCGATTGCCGGCTTTCCCGGAAGTACCGAGTATTAATCGCACGTGGACAGCTGACTCGTCAAATTATGGTACATGGCAACGTGTTACTGGCACAAAAGTGTCAATTTACTCGGCGTTCTATGACAATCGTACATCTCAGCCCTACATTCGTATCCTGGCCTTTTTTCACGGTCGCAACGCTTCAGCTGATCAAACACTATTCTGCCAAACGAGGTCACTCGACCCAAACGAAAATTCGGTAGAAGTTGTCGCAGCGAAACCGTTGGAAATTTGGTGGCACGAATGGGATGTTCAATCAACTGATATCGACTATCCATTTCTGCTGTCATGTCCATTAACAGGACCACTAAATGGACCCACATTAGTTTCACTAGTTACGGAGCCGTGTCTTGATCCCACAAATGCGCACCCGCTAAAACATAGTCTGAAAAGGAATCAACAAAAAAGGATGTTCACAATATGTGTTAAAGATTTGAATTTTGATAAGGATATCTCGCAAAATCTAATAGAATGGATAGAAACCAATAAAATACTCGGTGTTGACATTATCGATGTATACATCGGTAATATCAGTAAAGAAATAGAAAGTATCTTACAACACTATCGCGACGCGGGATTCATTAGATTGTTCCATGTTCCTATCAGAAACAAACCTGAAAGATCTCTATGGCAACGAAGGAGAGATCATATCATAACTTACAATGACTGTTTATATAGAAACATACAAGAAAGTGAATTTATTATACCATTAGATATCGATGAAATAGTTATGCCTAAGACCGCAAACTCTTTGCCTGAGTTATTAAAACATTTAGTCAAACGAGGTTGGAATCCTAAAGAATATTCCGCAATTATGATCAAAAACGTTTTCTTTTTTAAATTTATGCAAAAAATAAATAAGAATTATTATAATTATAAAAATAAGAGCAAAACATATGCAAAAAGGGATGATGTCCGAATAAGATATGCAAGGAGCACCAAAATCCTAGATGAAAACAATTCAGTACATTTGTTTGATAAAAATAAATTAGGTAATAAAAATTCAGTAGATGTGAACGAACTACAGGTATATTATGAAAATTACAATTCCAGATGCGGCCACGACATGCCGACGTCTAAATTAATGACTCATGTAATAAGTTCAGCATTCATTAGTCCTATAGGATTTTATAGTAAAAGTTTTATGGTCACCGAAAGAGTATTAACAGCCTTCAATCATTACCCTCTTGCTAGTCTTGGAACTTCAGGATTCGACGGTTGGCCTGCACCATTCAAAGAAGTTCAACTTAATCATTATAAGGAGTCGTGCAACGCGACCGTCGTGCCGGAGTGCGAGCTGTATCAACGCGACGTCCGCCTCGATCGAACCGCGCTGCGATTGAGAAAAAAACTGATCGAAGCACTATCGAATGCCATCTGTACATAA
Protein
MRTIIAYSQDIQEEMDRPSPVRCDRLPAFPEVPSINRTWTADSSNYGTWQRVTGTKVSIYSAFYDNRTSQPYIRILAFFHGRNASADQTLFCQTRSLDPNENSVEVVAAKPLEIWWHEWDVQSTDIDYPFLLSCPLTGPLNGPTLVSLVTEPCLDPTNAHPLKHSLKRNQQKRMFTICVKDLNFDKDISQNLIEWIETNKILGVDIIDVYIGNISKEIESILQHYRDAGFIRLFHVPIRNKPERSLWQRRRDHIITYNDCLYRNIQESEFIIPLDIDEIVMPKTANSLPELLKHLVKRGWNPKEYSAIMIKNVFFFKFMQKINKNYYNYKNKSKTYAKRDDVRIRYARSTKILDENNSVHLFDKNKLGNKNSVDVNELQVYYENYNSRCGHDMPTSKLMTHVISSAFISPIGFYSKSFMVTERVLTAFNHYPLASLGTSGFDGWPAPFKEVQLNHYKESCNATVVPECELYQRDVRLDRTALRLRKKLIEALSNAICT
Summary
Uniprot
A0A2H1UZW4
A0A3S2NPV9
A0A2W1C0A6
A0A212F463
A0A2A4J1V0
A0A194PTC1
+ More
A0A194QSL9 A0A0L7KE30 A0A2J7PXK0 A0A0L7QPC8 A0A088ACV8 A0A2A3ETD6 E2A315 A0A310SUW8 A0A0V1EU03 A0A0V0YBX5 A0A0V1G691 E2C7C8 A0A0V1JT83 A0A0V1I3F0 A0A195DCD8 F4WM62 T1HIZ0 A0A195BD98 A0A158P3F4 E2BH71 A0A195FUN5 A0A0L7RCA7 A0A151IGF0 A0A026WCB8 A0A023F4D9 E2AQ10 A0A0V1N0Q9 A0A0M8ZVX9 A0A2A3ESN5 V9ILB0 A0A087ZNN9 A0A310SF26 A0A151WFI4 A0A0V1NG95 A0A2S2PRH8 A0A0V0SMB8 A0A067RD35 A0A154P7X9 A0A1Y3EQS5 A0A0V1BXK9 A0A1Y3BLV5 A0A0V1CAZ6 A0A0V0VZT0 A0A0V1KUY1 A0A0P4VI58 A0A0V0TKD5 A0A0V1A1J5 A0A0V0X7H6 A0A3L8DYH6 A0A026VZ83 A0A0K2USQ1 A0A154NYI9 A0A0N5CUU4 A0A2R7VUM0 A0A132AC22 A0A1J1J168
A0A194QSL9 A0A0L7KE30 A0A2J7PXK0 A0A0L7QPC8 A0A088ACV8 A0A2A3ETD6 E2A315 A0A310SUW8 A0A0V1EU03 A0A0V0YBX5 A0A0V1G691 E2C7C8 A0A0V1JT83 A0A0V1I3F0 A0A195DCD8 F4WM62 T1HIZ0 A0A195BD98 A0A158P3F4 E2BH71 A0A195FUN5 A0A0L7RCA7 A0A151IGF0 A0A026WCB8 A0A023F4D9 E2AQ10 A0A0V1N0Q9 A0A0M8ZVX9 A0A2A3ESN5 V9ILB0 A0A087ZNN9 A0A310SF26 A0A151WFI4 A0A0V1NG95 A0A2S2PRH8 A0A0V0SMB8 A0A067RD35 A0A154P7X9 A0A1Y3EQS5 A0A0V1BXK9 A0A1Y3BLV5 A0A0V1CAZ6 A0A0V0VZT0 A0A0V1KUY1 A0A0P4VI58 A0A0V0TKD5 A0A0V1A1J5 A0A0V0X7H6 A0A3L8DYH6 A0A026VZ83 A0A0K2USQ1 A0A154NYI9 A0A0N5CUU4 A0A2R7VUM0 A0A132AC22 A0A1J1J168
Pubmed
EMBL
ODYU01000039
SOQ34140.1
RSAL01000163
RVE45351.1
KZ149904
PZC78406.1
+ More
AGBW02010434 OWR48521.1 NWSH01004152 PCG65420.1 KQ459599 KPI94380.1 KQ461154 KPJ08482.1 JTDY01010077 KOB61340.1 NEVH01020858 PNF21046.1 KQ414830 KOC60409.1 KZ288185 PBC34950.1 GL436262 EFN72173.1 KQ759841 OAD62629.1 JYDR01000011 JYDS01000023 KRY76499.1 KRZ31598.1 JYDU01000027 KRX97773.1 JYDT01000001 KRY93653.1 GL453369 EFN76115.1 JYDV01000049 KRZ38120.1 JYDP01000007 KRZ17433.1 KQ980989 KYN10575.1 GL888217 EGI64706.1 ACPB03006498 KQ976519 KYM82182.1 ADTU01001324 GL448268 EFN84992.1 KQ981264 KYN44002.1 KQ414616 KOC68468.1 KQ977726 KYN00304.1 KK107293 QOIP01000005 EZA53321.1 RLU22947.1 GBBI01002322 JAC16390.1 GL441645 EFN64497.1 JYDO01000017 KRZ77638.1 KQ435847 KOX71126.1 KZ288186 PBC34765.1 JR049744 AEY61059.1 KQ769179 OAD52905.1 KQ983219 KYQ46545.1 JYDM01000234 KRZ83017.1 GGMR01019404 MBY32023.1 JYDL01000003 KRX27589.1 KK852715 KDR17871.1 KQ434839 KZC08025.1 LVZM01007688 OUC46107.1 JYDH01000006 KRY41823.1 MUJZ01010920 OTF81959.1 JYDI01000298 KRY46264.1 JYDN01000001 KRX69003.1 JYDW01000238 KRZ51128.1 GDKW01002166 JAI54429.1 JYDJ01000235 KRX39392.1 JYDQ01000045 KRY18605.1 JYDK01000009 KRX83974.1 QOIP01000002 RLU25520.1 KK107544 EZA49077.1 HACA01023942 CDW41303.1 KQ434777 KZC04144.1 UYYF01004274 VDN01093.1 KK854054 PTY10390.1 JXLN01012465 KPM08518.1 CVRI01000064 CRL05516.1
AGBW02010434 OWR48521.1 NWSH01004152 PCG65420.1 KQ459599 KPI94380.1 KQ461154 KPJ08482.1 JTDY01010077 KOB61340.1 NEVH01020858 PNF21046.1 KQ414830 KOC60409.1 KZ288185 PBC34950.1 GL436262 EFN72173.1 KQ759841 OAD62629.1 JYDR01000011 JYDS01000023 KRY76499.1 KRZ31598.1 JYDU01000027 KRX97773.1 JYDT01000001 KRY93653.1 GL453369 EFN76115.1 JYDV01000049 KRZ38120.1 JYDP01000007 KRZ17433.1 KQ980989 KYN10575.1 GL888217 EGI64706.1 ACPB03006498 KQ976519 KYM82182.1 ADTU01001324 GL448268 EFN84992.1 KQ981264 KYN44002.1 KQ414616 KOC68468.1 KQ977726 KYN00304.1 KK107293 QOIP01000005 EZA53321.1 RLU22947.1 GBBI01002322 JAC16390.1 GL441645 EFN64497.1 JYDO01000017 KRZ77638.1 KQ435847 KOX71126.1 KZ288186 PBC34765.1 JR049744 AEY61059.1 KQ769179 OAD52905.1 KQ983219 KYQ46545.1 JYDM01000234 KRZ83017.1 GGMR01019404 MBY32023.1 JYDL01000003 KRX27589.1 KK852715 KDR17871.1 KQ434839 KZC08025.1 LVZM01007688 OUC46107.1 JYDH01000006 KRY41823.1 MUJZ01010920 OTF81959.1 JYDI01000298 KRY46264.1 JYDN01000001 KRX69003.1 JYDW01000238 KRZ51128.1 GDKW01002166 JAI54429.1 JYDJ01000235 KRX39392.1 JYDQ01000045 KRY18605.1 JYDK01000009 KRX83974.1 QOIP01000002 RLU25520.1 KK107544 EZA49077.1 HACA01023942 CDW41303.1 KQ434777 KZC04144.1 UYYF01004274 VDN01093.1 KK854054 PTY10390.1 JXLN01012465 KPM08518.1 CVRI01000064 CRL05516.1
Proteomes
UP000283053
UP000007151
UP000218220
UP000053268
UP000053240
UP000037510
+ More
UP000235965 UP000053825 UP000005203 UP000242457 UP000000311 UP000054632 UP000054805 UP000054815 UP000054995 UP000008237 UP000054826 UP000055024 UP000078492 UP000007755 UP000015103 UP000078540 UP000005205 UP000078541 UP000078542 UP000053097 UP000279307 UP000054843 UP000053105 UP000075809 UP000054924 UP000054630 UP000027135 UP000076502 UP000243006 UP000054776 UP000054653 UP000054681 UP000054721 UP000055048 UP000054783 UP000054673 UP000046394 UP000276776 UP000183832
UP000235965 UP000053825 UP000005203 UP000242457 UP000000311 UP000054632 UP000054805 UP000054815 UP000054995 UP000008237 UP000054826 UP000055024 UP000078492 UP000007755 UP000015103 UP000078540 UP000005205 UP000078541 UP000078542 UP000053097 UP000279307 UP000054843 UP000053105 UP000075809 UP000054924 UP000054630 UP000027135 UP000076502 UP000243006 UP000054776 UP000054653 UP000054681 UP000054721 UP000055048 UP000054783 UP000054673 UP000046394 UP000276776 UP000183832
Pfam
PF01697 Glyco_transf_92
Interpro
IPR008166
Glyco_transf_92
ProteinModelPortal
A0A2H1UZW4
A0A3S2NPV9
A0A2W1C0A6
A0A212F463
A0A2A4J1V0
A0A194PTC1
+ More
A0A194QSL9 A0A0L7KE30 A0A2J7PXK0 A0A0L7QPC8 A0A088ACV8 A0A2A3ETD6 E2A315 A0A310SUW8 A0A0V1EU03 A0A0V0YBX5 A0A0V1G691 E2C7C8 A0A0V1JT83 A0A0V1I3F0 A0A195DCD8 F4WM62 T1HIZ0 A0A195BD98 A0A158P3F4 E2BH71 A0A195FUN5 A0A0L7RCA7 A0A151IGF0 A0A026WCB8 A0A023F4D9 E2AQ10 A0A0V1N0Q9 A0A0M8ZVX9 A0A2A3ESN5 V9ILB0 A0A087ZNN9 A0A310SF26 A0A151WFI4 A0A0V1NG95 A0A2S2PRH8 A0A0V0SMB8 A0A067RD35 A0A154P7X9 A0A1Y3EQS5 A0A0V1BXK9 A0A1Y3BLV5 A0A0V1CAZ6 A0A0V0VZT0 A0A0V1KUY1 A0A0P4VI58 A0A0V0TKD5 A0A0V1A1J5 A0A0V0X7H6 A0A3L8DYH6 A0A026VZ83 A0A0K2USQ1 A0A154NYI9 A0A0N5CUU4 A0A2R7VUM0 A0A132AC22 A0A1J1J168
A0A194QSL9 A0A0L7KE30 A0A2J7PXK0 A0A0L7QPC8 A0A088ACV8 A0A2A3ETD6 E2A315 A0A310SUW8 A0A0V1EU03 A0A0V0YBX5 A0A0V1G691 E2C7C8 A0A0V1JT83 A0A0V1I3F0 A0A195DCD8 F4WM62 T1HIZ0 A0A195BD98 A0A158P3F4 E2BH71 A0A195FUN5 A0A0L7RCA7 A0A151IGF0 A0A026WCB8 A0A023F4D9 E2AQ10 A0A0V1N0Q9 A0A0M8ZVX9 A0A2A3ESN5 V9ILB0 A0A087ZNN9 A0A310SF26 A0A151WFI4 A0A0V1NG95 A0A2S2PRH8 A0A0V0SMB8 A0A067RD35 A0A154P7X9 A0A1Y3EQS5 A0A0V1BXK9 A0A1Y3BLV5 A0A0V1CAZ6 A0A0V0VZT0 A0A0V1KUY1 A0A0P4VI58 A0A0V0TKD5 A0A0V1A1J5 A0A0V0X7H6 A0A3L8DYH6 A0A026VZ83 A0A0K2USQ1 A0A154NYI9 A0A0N5CUU4 A0A2R7VUM0 A0A132AC22 A0A1J1J168
Ontologies
KEGG
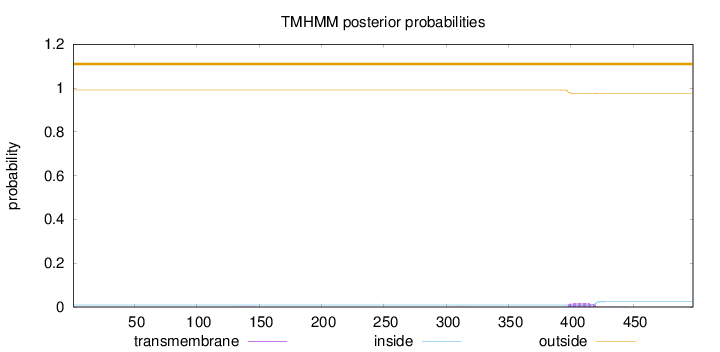
Topology
Length:
498
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.38275
Exp number, first 60 AAs:
0.00056
Total prob of N-in:
0.00928
outside
1 - 498
Population Genetic Test Statistics
Pi
271.246073
Theta
179.773563
Tajima's D
1.936358
CLR
0.137878
CSRT
0.870106494675266
Interpretation
Uncertain
