Gene
KWMTBOMO04556
Annotation
putative_rab-like_protein_5-like_isoform_1_[Danaus_plexippus]
Full name
Intraflagellar transport protein 22 homolog
Alternative Name
Rab-like protein 5
Location in the cell
Nuclear Reliability : 1.921
Sequence
CDS
ATGCAACCTAATAAATTGAAGATACTGATGATAGGTCCCTCTGAAAGTGGCAAAACTCAAATAGCTAATTTTTTATCGAATTCTATTAATATTGATGAAGCGGGAAACCCGAAGCCTACTCAAGGATTAAGAATAGTGGAATTCGAACAGTCAAATATAAACTATACCGGAAGAAACGTAGATATAGAATTGTGGGACTGTAGTGGTGATCATAGATTCGAATCATGCTGGCCTGCTTTAAGATTTGGAGTTAAAGGAGTAATTTTAGTTTGTTCATCTAACACATCAGAAGTCGCTCCTAGAGAATTAGAATTGCTATATAACTACTTTGTATCTCAACCAAAGCTATCCACTAAACAGTGTGTGGTGTTTTACAACTGCGTAGATGATCAATACGATGTTGGTCATCTTAATTTATCTTCGACATTTTCAAGAATATCTCAAGTCGCTGTGAATTTGAGAACAGAACGTGAACGTTTGAAAACTGAATTTTCTAATTACATAACCTCTGTACTTCAGTCGGTATAA
Protein
MQPNKLKILMIGPSESGKTQIANFLSNSINIDEAGNPKPTQGLRIVEFEQSNINYTGRNVDIELWDCSGDHRFESCWPALRFGVKGVILVCSSNTSEVAPRELELLYNYFVSQPKLSTKQCVVFYNCVDDQYDVGHLNLSSTFSRISQVAVNLRTERERLKTEFSNYITSVLQSV
Summary
Description
Small GTPase-like component of the intraflagellar transport (IFT) complex B.
Subunit
Component of the IFT complex B, at least composed of IFT20, IFT22, HSPB11/IFT25, IFT27, IFT46, IFT52, TRAF3IP1/IFT54, IFT57, IFT74, IFT80, IFT81, and IFT88. Interacts with IFT88 (By similarity).
Component of the IFT complex B, at least composed of IFT20, IFT22, HSPB11/IFT25, IFT27, IFT46, IFT52, TRAF3IP1/IFT54, IFT57, IFT74, IFT80, IFT81, and IFT88. Interacts with IFT88.
Component of the IFT complex B, at least composed of IFT20, IFT22, HSPB11/IFT25, IFT27, IFT46, IFT52, TRAF3IP1/IFT54, IFT57, IFT74, IFT80, IFT81, and IFT88. Interacts with IFT88.
Similarity
Belongs to the small GTPase superfamily. Rab family.
Keywords
Alternative splicing
Cell projection
Cilium
Complete proteome
GTP-binding
Nucleotide-binding
Phosphoprotein
Reference proteome
Feature
chain Intraflagellar transport protein 22 homolog
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
A0A2W1BXN1
A0A2H1UZZ6
A0A212F476
A0A0L7LUD8
A0A067R4E3
A0A087ZUD6
+ More
A0A0L7QNU4 W4Z5V1 A0A154PLM0 E2BI81 A0A3L8DG41 A0A3R7SPR1 E2AXA6 A0A3B3TES6 V9KJA2 A0A2R9ADG8 H2QV55 A0A024QYV3 Q9H7X7 A0A2U3VUQ0 A0A1B6CYT5 A0A2J8XU70 A0A026VZP2 A0A2Y9I237 A0A2U3XQT6 A0A1B6F5E3 A0A3Q7NPS1 A0A1S3GEF7 T2M5X3 A0A2K5IMY4 A0A250XVW4 A0A1B6IK14 U3EK23 A0A0D9RZL5 Q4R4K5 G5AXB6 A0A2K6Q8G0 R7UG98 F6PS53 A0A2A3EJL7 A0A2K6EN03 A0A3Q7XBR6 I3MIL2 A0A151X8C9 A0A2K5NXF2 A0A2K6TR13 G1PKZ0 A0A337SN32 A0A2K5ZPZ0 A0A158NP14 A0A096NR58 G3U3H5 M3YWV4 A0A2K5QW05 F7A8A4 A0A0P4WF57 A0A2Y9J931 A0A3Q0CGZ2 G1STM1 A0A087UQT1 H0WFM3 S4RWD7 A0A1W4XA72 A0A3Q7UBY9 A0A0L8HD41 F6V8Z0 A7S8S6 F1RKG2 H2YA04 A0A1S3H1D7 A0A384AHF1 A0A2A2KQW2 H3BFQ7 Q9DAI2 F7CN84 A0A2Y9FLW5 Q5FVJ7 C1BW53 A0A3B1IVU6 A0A1A6GH16 A0A1S2ZE45 A0A2U3UZM3 A0A341BHL5 A0A091DWJ0 A0A2Y9PLW1 L8IQU7 A0A3P9KB75 A0A286XRH9 A0A147BPA0 B5X756 A0A0P7WDB2 A0A340Y6M1 E3TDW6 A0A2G9U682 A0A1W4Z174 A0A3Q2PQZ4 W5PZK3 A0A3Q1GDG7 A0A3Q1B443 A0A3P8UHQ8
A0A0L7QNU4 W4Z5V1 A0A154PLM0 E2BI81 A0A3L8DG41 A0A3R7SPR1 E2AXA6 A0A3B3TES6 V9KJA2 A0A2R9ADG8 H2QV55 A0A024QYV3 Q9H7X7 A0A2U3VUQ0 A0A1B6CYT5 A0A2J8XU70 A0A026VZP2 A0A2Y9I237 A0A2U3XQT6 A0A1B6F5E3 A0A3Q7NPS1 A0A1S3GEF7 T2M5X3 A0A2K5IMY4 A0A250XVW4 A0A1B6IK14 U3EK23 A0A0D9RZL5 Q4R4K5 G5AXB6 A0A2K6Q8G0 R7UG98 F6PS53 A0A2A3EJL7 A0A2K6EN03 A0A3Q7XBR6 I3MIL2 A0A151X8C9 A0A2K5NXF2 A0A2K6TR13 G1PKZ0 A0A337SN32 A0A2K5ZPZ0 A0A158NP14 A0A096NR58 G3U3H5 M3YWV4 A0A2K5QW05 F7A8A4 A0A0P4WF57 A0A2Y9J931 A0A3Q0CGZ2 G1STM1 A0A087UQT1 H0WFM3 S4RWD7 A0A1W4XA72 A0A3Q7UBY9 A0A0L8HD41 F6V8Z0 A7S8S6 F1RKG2 H2YA04 A0A1S3H1D7 A0A384AHF1 A0A2A2KQW2 H3BFQ7 Q9DAI2 F7CN84 A0A2Y9FLW5 Q5FVJ7 C1BW53 A0A3B1IVU6 A0A1A6GH16 A0A1S2ZE45 A0A2U3UZM3 A0A341BHL5 A0A091DWJ0 A0A2Y9PLW1 L8IQU7 A0A3P9KB75 A0A286XRH9 A0A147BPA0 B5X756 A0A0P7WDB2 A0A340Y6M1 E3TDW6 A0A2G9U682 A0A1W4Z174 A0A3Q2PQZ4 W5PZK3 A0A3Q1GDG7 A0A3Q1B443 A0A3P8UHQ8
Pubmed
28756777
22118469
26227816
24845553
20798317
30249741
+ More
29240929 24402279 22722832 16136131 11181995 14702039 12853948 15489334 17974005 23186163 24508170 24065732 28087693 25243066 21993625 25362486 23254933 17431167 25319552 21993624 17975172 21347285 19892987 28071753 16341006 17615350 9215903 16141072 19253336 21183079 17495919 20433749 25329095 22751099 17554307 29652888 20634964 20809919
29240929 24402279 22722832 16136131 11181995 14702039 12853948 15489334 17974005 23186163 24508170 24065732 28087693 25243066 21993625 25362486 23254933 17431167 25319552 21993624 17975172 21347285 19892987 28071753 16341006 17615350 9215903 16141072 19253336 21183079 17495919 20433749 25329095 22751099 17554307 29652888 20634964 20809919
EMBL
KZ149904
PZC78405.1
ODYU01000039
SOQ34139.1
AGBW02010434
OWR48522.1
+ More
JTDY01000068 KOB79070.1 KK852704 KDR18061.1 KQ414855 KOC60161.1 AAGJ04121967 AAGJ04121968 KQ434938 KZC12120.1 GL448457 EFN84612.1 QOIP01000008 RLU19374.1 QCYY01002508 ROT69876.1 GL443548 EFN61886.1 JW865663 AFO98180.1 AJFE02020806 AJFE02020807 AC187615 GABC01009638 GABD01007837 NBAG03000065 JAA01700.1 JAA25263.1 PNI92632.1 PNI92634.1 CH471197 EAW50218.1 AK023287 AK024179 AK298888 AC006329 BC004522 BC009823 BC038668 AL157469 GEDC01018813 JAS18485.1 ABGA01059864 ABGA01059865 NDHI03003310 PNJ85584.1 PNJ85586.1 KK107648 EZA48354.1 GECZ01024359 GECZ01009590 JAS45410.1 JAS60179.1 HAAD01001083 CDG67315.1 GFFV01004484 JAV35461.1 GECU01020447 JAS87259.1 GAMR01002526 GAMQ01004855 GAMP01010136 JAB31406.1 JAB36996.1 JAB42619.1 AQIB01139502 AB169889 JH167379 GEBF01006548 EHB01678.1 JAN97084.1 AMQN01008687 KB303737 ELU02828.1 JSUE03029184 JU335951 JU476832 JV047955 AFE79704.1 AFH33636.1 AFI38026.1 KZ288223 PBC32003.1 AGTP01053258 KQ982422 KYQ56623.1 AAPE02016753 AANG04004562 ADTU01022032 AHZZ02022087 AEYP01053164 GDRN01061587 GDRN01061586 JAI65195.1 KK121096 KFM79720.1 AAQR03108005 AAQR03108006 KQ418495 KOF87097.1 AAEX03004261 DS469599 EDO39919.1 AEMK02000015 LIAE01007929 PAV76207.1 AFYH01014644 AK005820 BC009150 BC089940 BT078832 ACO13256.1 LZPO01097164 OBS64582.1 KN121778 KFO35462.1 JH880833 ELR58438.1 AAKN02032072 GEGO01003092 JAR92312.1 BT046875 ACI66676.1 JARO02009956 KPP61127.1 GU588546 ADO28502.1 KZ348740 PIO65779.1 AMGL01069406
JTDY01000068 KOB79070.1 KK852704 KDR18061.1 KQ414855 KOC60161.1 AAGJ04121967 AAGJ04121968 KQ434938 KZC12120.1 GL448457 EFN84612.1 QOIP01000008 RLU19374.1 QCYY01002508 ROT69876.1 GL443548 EFN61886.1 JW865663 AFO98180.1 AJFE02020806 AJFE02020807 AC187615 GABC01009638 GABD01007837 NBAG03000065 JAA01700.1 JAA25263.1 PNI92632.1 PNI92634.1 CH471197 EAW50218.1 AK023287 AK024179 AK298888 AC006329 BC004522 BC009823 BC038668 AL157469 GEDC01018813 JAS18485.1 ABGA01059864 ABGA01059865 NDHI03003310 PNJ85584.1 PNJ85586.1 KK107648 EZA48354.1 GECZ01024359 GECZ01009590 JAS45410.1 JAS60179.1 HAAD01001083 CDG67315.1 GFFV01004484 JAV35461.1 GECU01020447 JAS87259.1 GAMR01002526 GAMQ01004855 GAMP01010136 JAB31406.1 JAB36996.1 JAB42619.1 AQIB01139502 AB169889 JH167379 GEBF01006548 EHB01678.1 JAN97084.1 AMQN01008687 KB303737 ELU02828.1 JSUE03029184 JU335951 JU476832 JV047955 AFE79704.1 AFH33636.1 AFI38026.1 KZ288223 PBC32003.1 AGTP01053258 KQ982422 KYQ56623.1 AAPE02016753 AANG04004562 ADTU01022032 AHZZ02022087 AEYP01053164 GDRN01061587 GDRN01061586 JAI65195.1 KK121096 KFM79720.1 AAQR03108005 AAQR03108006 KQ418495 KOF87097.1 AAEX03004261 DS469599 EDO39919.1 AEMK02000015 LIAE01007929 PAV76207.1 AFYH01014644 AK005820 BC009150 BC089940 BT078832 ACO13256.1 LZPO01097164 OBS64582.1 KN121778 KFO35462.1 JH880833 ELR58438.1 AAKN02032072 GEGO01003092 JAR92312.1 BT046875 ACI66676.1 JARO02009956 KPP61127.1 GU588546 ADO28502.1 KZ348740 PIO65779.1 AMGL01069406
Proteomes
UP000007151
UP000037510
UP000027135
UP000005203
UP000053825
UP000007110
+ More
UP000076502 UP000008237 UP000279307 UP000283509 UP000000311 UP000261540 UP000240080 UP000002277 UP000005640 UP000245340 UP000001595 UP000053097 UP000248481 UP000245341 UP000286641 UP000081671 UP000233080 UP000008225 UP000029965 UP000233100 UP000006813 UP000233200 UP000014760 UP000006718 UP000242457 UP000233160 UP000286642 UP000005215 UP000075809 UP000233060 UP000233220 UP000001074 UP000011712 UP000233140 UP000005205 UP000028761 UP000007646 UP000000715 UP000233040 UP000002281 UP000248482 UP000189706 UP000001811 UP000054359 UP000005225 UP000245300 UP000192223 UP000286640 UP000053454 UP000002254 UP000001593 UP000008227 UP000007875 UP000085678 UP000261681 UP000218231 UP000008672 UP000000589 UP000002280 UP000248484 UP000002494 UP000018467 UP000092124 UP000079721 UP000245320 UP000252040 UP000028990 UP000248483 UP000265180 UP000005447 UP000087266 UP000034805 UP000265300 UP000221080 UP000192224 UP000265000 UP000002356 UP000257200 UP000257160 UP000265080
UP000076502 UP000008237 UP000279307 UP000283509 UP000000311 UP000261540 UP000240080 UP000002277 UP000005640 UP000245340 UP000001595 UP000053097 UP000248481 UP000245341 UP000286641 UP000081671 UP000233080 UP000008225 UP000029965 UP000233100 UP000006813 UP000233200 UP000014760 UP000006718 UP000242457 UP000233160 UP000286642 UP000005215 UP000075809 UP000233060 UP000233220 UP000001074 UP000011712 UP000233140 UP000005205 UP000028761 UP000007646 UP000000715 UP000233040 UP000002281 UP000248482 UP000189706 UP000001811 UP000054359 UP000005225 UP000245300 UP000192223 UP000286640 UP000053454 UP000002254 UP000001593 UP000008227 UP000007875 UP000085678 UP000261681 UP000218231 UP000008672 UP000000589 UP000002280 UP000248484 UP000002494 UP000018467 UP000092124 UP000079721 UP000245320 UP000252040 UP000028990 UP000248483 UP000265180 UP000005447 UP000087266 UP000034805 UP000265300 UP000221080 UP000192224 UP000265000 UP000002356 UP000257200 UP000257160 UP000265080
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
A0A2W1BXN1
A0A2H1UZZ6
A0A212F476
A0A0L7LUD8
A0A067R4E3
A0A087ZUD6
+ More
A0A0L7QNU4 W4Z5V1 A0A154PLM0 E2BI81 A0A3L8DG41 A0A3R7SPR1 E2AXA6 A0A3B3TES6 V9KJA2 A0A2R9ADG8 H2QV55 A0A024QYV3 Q9H7X7 A0A2U3VUQ0 A0A1B6CYT5 A0A2J8XU70 A0A026VZP2 A0A2Y9I237 A0A2U3XQT6 A0A1B6F5E3 A0A3Q7NPS1 A0A1S3GEF7 T2M5X3 A0A2K5IMY4 A0A250XVW4 A0A1B6IK14 U3EK23 A0A0D9RZL5 Q4R4K5 G5AXB6 A0A2K6Q8G0 R7UG98 F6PS53 A0A2A3EJL7 A0A2K6EN03 A0A3Q7XBR6 I3MIL2 A0A151X8C9 A0A2K5NXF2 A0A2K6TR13 G1PKZ0 A0A337SN32 A0A2K5ZPZ0 A0A158NP14 A0A096NR58 G3U3H5 M3YWV4 A0A2K5QW05 F7A8A4 A0A0P4WF57 A0A2Y9J931 A0A3Q0CGZ2 G1STM1 A0A087UQT1 H0WFM3 S4RWD7 A0A1W4XA72 A0A3Q7UBY9 A0A0L8HD41 F6V8Z0 A7S8S6 F1RKG2 H2YA04 A0A1S3H1D7 A0A384AHF1 A0A2A2KQW2 H3BFQ7 Q9DAI2 F7CN84 A0A2Y9FLW5 Q5FVJ7 C1BW53 A0A3B1IVU6 A0A1A6GH16 A0A1S2ZE45 A0A2U3UZM3 A0A341BHL5 A0A091DWJ0 A0A2Y9PLW1 L8IQU7 A0A3P9KB75 A0A286XRH9 A0A147BPA0 B5X756 A0A0P7WDB2 A0A340Y6M1 E3TDW6 A0A2G9U682 A0A1W4Z174 A0A3Q2PQZ4 W5PZK3 A0A3Q1GDG7 A0A3Q1B443 A0A3P8UHQ8
A0A0L7QNU4 W4Z5V1 A0A154PLM0 E2BI81 A0A3L8DG41 A0A3R7SPR1 E2AXA6 A0A3B3TES6 V9KJA2 A0A2R9ADG8 H2QV55 A0A024QYV3 Q9H7X7 A0A2U3VUQ0 A0A1B6CYT5 A0A2J8XU70 A0A026VZP2 A0A2Y9I237 A0A2U3XQT6 A0A1B6F5E3 A0A3Q7NPS1 A0A1S3GEF7 T2M5X3 A0A2K5IMY4 A0A250XVW4 A0A1B6IK14 U3EK23 A0A0D9RZL5 Q4R4K5 G5AXB6 A0A2K6Q8G0 R7UG98 F6PS53 A0A2A3EJL7 A0A2K6EN03 A0A3Q7XBR6 I3MIL2 A0A151X8C9 A0A2K5NXF2 A0A2K6TR13 G1PKZ0 A0A337SN32 A0A2K5ZPZ0 A0A158NP14 A0A096NR58 G3U3H5 M3YWV4 A0A2K5QW05 F7A8A4 A0A0P4WF57 A0A2Y9J931 A0A3Q0CGZ2 G1STM1 A0A087UQT1 H0WFM3 S4RWD7 A0A1W4XA72 A0A3Q7UBY9 A0A0L8HD41 F6V8Z0 A7S8S6 F1RKG2 H2YA04 A0A1S3H1D7 A0A384AHF1 A0A2A2KQW2 H3BFQ7 Q9DAI2 F7CN84 A0A2Y9FLW5 Q5FVJ7 C1BW53 A0A3B1IVU6 A0A1A6GH16 A0A1S2ZE45 A0A2U3UZM3 A0A341BHL5 A0A091DWJ0 A0A2Y9PLW1 L8IQU7 A0A3P9KB75 A0A286XRH9 A0A147BPA0 B5X756 A0A0P7WDB2 A0A340Y6M1 E3TDW6 A0A2G9U682 A0A1W4Z174 A0A3Q2PQZ4 W5PZK3 A0A3Q1GDG7 A0A3Q1B443 A0A3P8UHQ8
PDB
6IAE
E-value=6.91376e-08,
Score=131
Ontologies
GO
Topology
Subcellular location
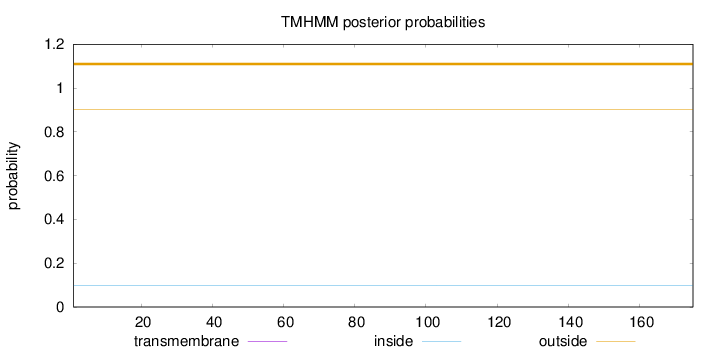
Length:
175
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00122
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09691
outside
1 - 175
Population Genetic Test Statistics
Pi
337.757962
Theta
207.966228
Tajima's D
2.012605
CLR
0.129886
CSRT
0.888355582220889
Interpretation
Uncertain
