Pre Gene Modal
BGIBMGA005469
Annotation
PREDICTED:_DNA-directed_RNA_polymerase_II_16_kDa_polypeptide_[Amyelois_transitella]
Full name
DNA-directed RNA polymerase II subunit Rpb4
Location in the cell
Cytoplasmic Reliability : 1.826 Nuclear Reliability : 2.233
Sequence
CDS
ATGACTGGACCTCTACAATATGTGGTGGAAGAGGATGCAGCTGATCTGCAATTCCCAAAAGAATTTGAGAATGCTGAAACTTTAATGATATCTGAAGTTGACATGTTATTGGAAAATAGAAAAGCTCAAAATGAATCTGCTGAAGAGGAACAAGAATTCTCTGAAGTCTTCATGAAAACACTAACGTATACCAATATGTTTAAAAAGTTCAAAAACAAAGAGACAATAGCTGCTGTTAGACACCTATTACAATCAAAAAAACTTCACAAGTTTGAATTGGCAAGCTTAGCAAATTTATGTCCTGAAACACCGGAGGAAGCGAAAGCACTGATACCATCATTAGAAGGAAAATTTGAAGATGAAGAACTAAGAATACTTTTGGATGACATACAAACTAAGAGAAGTTTACAATACTAA
Protein
MTGPLQYVVEEDAADLQFPKEFENAETLMISEVDMLLENRKAQNESAEEEQEFSEVFMKTLTYTNMFKKFKNKETIAAVRHLLQSKKLHKFELASLANLCPETPEEAKALIPSLEGKFEDEELRILLDDIQTKRSLQY
Summary
Description
DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Associates with POLR2G.
Subunit
RNA polymerase II consists of 12 different subunits.
Miscellaneous
This protein is produced by a bicistronic gene which also produces the Ada2a protein by alternative splicing (PubMed:19921261). Three distinct zinc-containing RNA polymerases are found in eukaryotic nuclei: polymerase I for the ribosomal RNA precursor, polymerase II for the mRNA precursor, and polymerase III for 5S and tRNA genes (Probable).
Similarity
Belongs to the eukaryotic RPB4 RNA polymerase subunit family.
Keywords
Alternative splicing
Chromosome
Complete proteome
DNA-directed RNA polymerase
Nucleus
Reference proteome
Transcription
Feature
chain DNA-directed RNA polymerase II subunit Rpb4
Uniprot
H9J7H7
A0A1E1WTC5
A0A2A4IYL1
A0A2H1UZV5
A0A212F456
S4PXV3
+ More
A0A0L7LKF0 A0A2W1BVN3 A0A194QSH8 I4DK70 A0A194PNR5 A0A2J7PJH7 K7JC40 A0A0C9R9N0 A0A1B6CJH2 A0A1B6ME97 A0A1B6FAU2 A0A0P4VY13 A0A069DVZ2 R4G820 A0A0V0G8N2 A0A023FAV0 A0A1B6IZR8 A0A1Y1L2V3 D6WIZ2 A0A2R7X5U9 A0A023EG90 A0A0K8TS37 A0A1S3DJH0 B0WK75 Q175I0 A0A1Q3FD64 W8BN20 A0A0K8UDC6 A0A0C9QL36 A0A0A1XT68 A0A1L8DGE0 A0A1I8N4B4 A0A1I8NLM1 N6TBU0 A0A0A9XSM1 U5ERW6 A0A1W4UK81 A0A0Q9WH43 A0A0Q9X0N4 A0A0R3NNP8 A0A0R1E0T3 A0A0P8XTR8 A0A336M5M8 A0A0P6DGL0 A0A0P5MMG7 A0A182NQR8 A0A182QJ50 A0A182IXN9 C3XPX8 A0A1B2AK30 Q9VEA5 E9FYU9 A0A0P4XCV5 A0A0Q9WWF1 A0A0P5H0X3 T1DH95 A0A2M4AW40 A0A2M4C2U6 W5J580 A0A182FS24 A0A2M3ZIT3 A0A182YHX8 A0A182PRU8 A0A182MSX9 A0A182SCG4 A0A182VKK5 A0A182K7I6 A0A182VW03 A0A182UAM8 A0A182X9D5 F5HMV9 A0A182RSC9 A0A182HKC1 A0A3S3PI72 A0A1J1IHK1 G3MMV2 A0A1E1XTZ6 A0A023FFE1 A0A131XG95 A0A1E1WXK1 A0A023GEV0 G3MMV1 A0A023FUH5 H3BCJ8 A0A087TQ00 A0A224YVY4 A0A131YF29 L7M2X9 T1J3Q6 K1QN72 Q6DRG4 A0A3N0YKF7 E3TD45 E3TGE1
A0A0L7LKF0 A0A2W1BVN3 A0A194QSH8 I4DK70 A0A194PNR5 A0A2J7PJH7 K7JC40 A0A0C9R9N0 A0A1B6CJH2 A0A1B6ME97 A0A1B6FAU2 A0A0P4VY13 A0A069DVZ2 R4G820 A0A0V0G8N2 A0A023FAV0 A0A1B6IZR8 A0A1Y1L2V3 D6WIZ2 A0A2R7X5U9 A0A023EG90 A0A0K8TS37 A0A1S3DJH0 B0WK75 Q175I0 A0A1Q3FD64 W8BN20 A0A0K8UDC6 A0A0C9QL36 A0A0A1XT68 A0A1L8DGE0 A0A1I8N4B4 A0A1I8NLM1 N6TBU0 A0A0A9XSM1 U5ERW6 A0A1W4UK81 A0A0Q9WH43 A0A0Q9X0N4 A0A0R3NNP8 A0A0R1E0T3 A0A0P8XTR8 A0A336M5M8 A0A0P6DGL0 A0A0P5MMG7 A0A182NQR8 A0A182QJ50 A0A182IXN9 C3XPX8 A0A1B2AK30 Q9VEA5 E9FYU9 A0A0P4XCV5 A0A0Q9WWF1 A0A0P5H0X3 T1DH95 A0A2M4AW40 A0A2M4C2U6 W5J580 A0A182FS24 A0A2M3ZIT3 A0A182YHX8 A0A182PRU8 A0A182MSX9 A0A182SCG4 A0A182VKK5 A0A182K7I6 A0A182VW03 A0A182UAM8 A0A182X9D5 F5HMV9 A0A182RSC9 A0A182HKC1 A0A3S3PI72 A0A1J1IHK1 G3MMV2 A0A1E1XTZ6 A0A023FFE1 A0A131XG95 A0A1E1WXK1 A0A023GEV0 G3MMV1 A0A023FUH5 H3BCJ8 A0A087TQ00 A0A224YVY4 A0A131YF29 L7M2X9 T1J3Q6 K1QN72 Q6DRG4 A0A3N0YKF7 E3TD45 E3TGE1
Pubmed
19121390
22118469
23622113
26227816
28756777
26354079
+ More
22651552 20075255 27129103 26334808 25474469 28004739 18362917 19820115 24945155 26369729 17510324 24495485 25830018 25315136 23537049 25401762 26823975 17994087 15632085 17550304 18563158 12482983 10731132 12537572 12697829 19921261 21292972 20920257 23761445 25244985 12364791 14747013 17210077 22216098 29209593 28049606 28503490 9215903 28797301 26830274 25576852 22992520 15256591 23594743 20634964
22651552 20075255 27129103 26334808 25474469 28004739 18362917 19820115 24945155 26369729 17510324 24495485 25830018 25315136 23537049 25401762 26823975 17994087 15632085 17550304 18563158 12482983 10731132 12537572 12697829 19921261 21292972 20920257 23761445 25244985 12364791 14747013 17210077 22216098 29209593 28049606 28503490 9215903 28797301 26830274 25576852 22992520 15256591 23594743 20634964
EMBL
BABH01013858
GDQN01000804
JAT90250.1
NWSH01004628
PCG64841.1
ODYU01000039
+ More
SOQ34135.1 AGBW02010434 OWR48526.1 GAIX01004388 JAA88172.1 JTDY01000778 KOB75910.1 KZ149904 PZC78401.1 KQ461154 KPJ08478.1 AK401688 BAM18310.1 KQ459599 KPI94384.1 NEVH01024949 PNF16469.1 AAZX01017896 GBYB01003496 JAG73263.1 GEDC01023823 JAS13475.1 GEBQ01005747 GEBQ01004200 JAT34230.1 JAT35777.1 GECZ01022399 GECZ01016462 JAS47370.1 JAS53307.1 GDKW01002118 JAI54477.1 GBGD01003415 JAC85474.1 ACPB03000895 GAHY01001737 JAA75773.1 GECL01002000 JAP04124.1 GBBI01000553 JAC18159.1 GECU01036241 GECU01015262 JAS71465.1 JAS92444.1 GEZM01068928 JAV66700.1 KQ971342 EFA04771.1 KK857435 PTY27168.1 GAPW01005758 GEHC01001245 JAC07840.1 JAV46400.1 GDAI01000863 JAI16740.1 DS231968 EDS29649.1 CH477400 EAT41740.1 GFDL01009586 JAV25459.1 GAMC01011784 JAB94771.1 GDHF01027949 JAI24365.1 GBYB01004244 JAG74011.1 GBXI01000036 JAD14256.1 GFDF01008562 JAV05522.1 APGK01043807 KB741018 ENN75193.1 GBHO01019882 GBRD01015398 GDHC01015475 JAG23722.1 JAG50428.1 JAQ03154.1 GANO01002642 JAB57229.1 CH940650 KRF83832.1 CH933806 KRG01517.1 CM000070 KRT00659.1 CM000160 KRK02777.1 CH902623 KPU72722.1 UFQT01000590 SSX25545.1 GDIQ01080867 JAN13870.1 GDIQ01165494 JAK86231.1 AXCN02001501 GG666451 EEN69999.1 KX531723 ANY27533.1 AF544019 AF544020 AE014297 BT022168 BT023308 BT023309 AAN88031.1 AAY51562.1 GL732527 EFX87599.1 GDIP01244922 JAI78479.1 CH964251 KRF99768.1 GDIQ01256281 JAJ95443.1 GAMD01002456 JAA99134.1 GGFK01011507 MBW44828.1 GGFJ01010505 MBW59646.1 ADMH02002101 GGFL01004241 ETN59136.1 MBW68419.1 GGFM01007705 MBW28456.1 AXCM01001239 AAAB01008987 EGK97631.1 APCN01000825 NCKU01002503 NCKU01002219 RWS09440.1 RWS10116.1 CVRI01000052 CRK99699.1 JO843203 AEO34820.1 GFAA01000639 JAU02796.1 GBBK01004195 GBBK01004192 JAC20287.1 GEFH01003850 JAP64731.1 GFAC01007481 JAT91707.1 GBBM01002991 JAC32427.1 JO843202 AEO34819.1 GBBL01002236 JAC25084.1 AFYH01032347 AFYH01032348 KK116251 KFM67189.1 GFPF01007317 MAA18463.1 GEDV01011040 JAP77517.1 GACK01006368 JAA58666.1 JH431830 JH818600 EKC30315.1 CU638696 BC095261 BC164278 AY648795 AAH95261.1 AAI64278.1 AAT68113.1 RJVU01037189 ROL46745.1 GU588275 ADO28231.1 GU589427 ADO29377.1
SOQ34135.1 AGBW02010434 OWR48526.1 GAIX01004388 JAA88172.1 JTDY01000778 KOB75910.1 KZ149904 PZC78401.1 KQ461154 KPJ08478.1 AK401688 BAM18310.1 KQ459599 KPI94384.1 NEVH01024949 PNF16469.1 AAZX01017896 GBYB01003496 JAG73263.1 GEDC01023823 JAS13475.1 GEBQ01005747 GEBQ01004200 JAT34230.1 JAT35777.1 GECZ01022399 GECZ01016462 JAS47370.1 JAS53307.1 GDKW01002118 JAI54477.1 GBGD01003415 JAC85474.1 ACPB03000895 GAHY01001737 JAA75773.1 GECL01002000 JAP04124.1 GBBI01000553 JAC18159.1 GECU01036241 GECU01015262 JAS71465.1 JAS92444.1 GEZM01068928 JAV66700.1 KQ971342 EFA04771.1 KK857435 PTY27168.1 GAPW01005758 GEHC01001245 JAC07840.1 JAV46400.1 GDAI01000863 JAI16740.1 DS231968 EDS29649.1 CH477400 EAT41740.1 GFDL01009586 JAV25459.1 GAMC01011784 JAB94771.1 GDHF01027949 JAI24365.1 GBYB01004244 JAG74011.1 GBXI01000036 JAD14256.1 GFDF01008562 JAV05522.1 APGK01043807 KB741018 ENN75193.1 GBHO01019882 GBRD01015398 GDHC01015475 JAG23722.1 JAG50428.1 JAQ03154.1 GANO01002642 JAB57229.1 CH940650 KRF83832.1 CH933806 KRG01517.1 CM000070 KRT00659.1 CM000160 KRK02777.1 CH902623 KPU72722.1 UFQT01000590 SSX25545.1 GDIQ01080867 JAN13870.1 GDIQ01165494 JAK86231.1 AXCN02001501 GG666451 EEN69999.1 KX531723 ANY27533.1 AF544019 AF544020 AE014297 BT022168 BT023308 BT023309 AAN88031.1 AAY51562.1 GL732527 EFX87599.1 GDIP01244922 JAI78479.1 CH964251 KRF99768.1 GDIQ01256281 JAJ95443.1 GAMD01002456 JAA99134.1 GGFK01011507 MBW44828.1 GGFJ01010505 MBW59646.1 ADMH02002101 GGFL01004241 ETN59136.1 MBW68419.1 GGFM01007705 MBW28456.1 AXCM01001239 AAAB01008987 EGK97631.1 APCN01000825 NCKU01002503 NCKU01002219 RWS09440.1 RWS10116.1 CVRI01000052 CRK99699.1 JO843203 AEO34820.1 GFAA01000639 JAU02796.1 GBBK01004195 GBBK01004192 JAC20287.1 GEFH01003850 JAP64731.1 GFAC01007481 JAT91707.1 GBBM01002991 JAC32427.1 JO843202 AEO34819.1 GBBL01002236 JAC25084.1 AFYH01032347 AFYH01032348 KK116251 KFM67189.1 GFPF01007317 MAA18463.1 GEDV01011040 JAP77517.1 GACK01006368 JAA58666.1 JH431830 JH818600 EKC30315.1 CU638696 BC095261 BC164278 AY648795 AAH95261.1 AAI64278.1 AAT68113.1 RJVU01037189 ROL46745.1 GU588275 ADO28231.1 GU589427 ADO29377.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000053240
UP000053268
+ More
UP000235965 UP000002358 UP000015103 UP000007266 UP000079169 UP000002320 UP000008820 UP000095301 UP000095300 UP000019118 UP000192221 UP000008792 UP000009192 UP000001819 UP000002282 UP000007801 UP000075884 UP000075886 UP000075880 UP000001554 UP000000803 UP000000305 UP000007798 UP000000673 UP000069272 UP000076408 UP000075885 UP000075883 UP000075901 UP000075903 UP000075881 UP000075920 UP000075902 UP000076407 UP000007062 UP000075900 UP000075840 UP000285301 UP000183832 UP000008672 UP000054359 UP000005408 UP000000437 UP000221080
UP000235965 UP000002358 UP000015103 UP000007266 UP000079169 UP000002320 UP000008820 UP000095301 UP000095300 UP000019118 UP000192221 UP000008792 UP000009192 UP000001819 UP000002282 UP000007801 UP000075884 UP000075886 UP000075880 UP000001554 UP000000803 UP000000305 UP000007798 UP000000673 UP000069272 UP000076408 UP000075885 UP000075883 UP000075901 UP000075903 UP000075881 UP000075920 UP000075902 UP000076407 UP000007062 UP000075900 UP000075840 UP000285301 UP000183832 UP000008672 UP000054359 UP000005408 UP000000437 UP000221080
Pfam
PF03874 RNA_pol_Rpb4
Interpro
SUPFAM
SSF47819
SSF47819
Gene 3D
ProteinModelPortal
H9J7H7
A0A1E1WTC5
A0A2A4IYL1
A0A2H1UZV5
A0A212F456
S4PXV3
+ More
A0A0L7LKF0 A0A2W1BVN3 A0A194QSH8 I4DK70 A0A194PNR5 A0A2J7PJH7 K7JC40 A0A0C9R9N0 A0A1B6CJH2 A0A1B6ME97 A0A1B6FAU2 A0A0P4VY13 A0A069DVZ2 R4G820 A0A0V0G8N2 A0A023FAV0 A0A1B6IZR8 A0A1Y1L2V3 D6WIZ2 A0A2R7X5U9 A0A023EG90 A0A0K8TS37 A0A1S3DJH0 B0WK75 Q175I0 A0A1Q3FD64 W8BN20 A0A0K8UDC6 A0A0C9QL36 A0A0A1XT68 A0A1L8DGE0 A0A1I8N4B4 A0A1I8NLM1 N6TBU0 A0A0A9XSM1 U5ERW6 A0A1W4UK81 A0A0Q9WH43 A0A0Q9X0N4 A0A0R3NNP8 A0A0R1E0T3 A0A0P8XTR8 A0A336M5M8 A0A0P6DGL0 A0A0P5MMG7 A0A182NQR8 A0A182QJ50 A0A182IXN9 C3XPX8 A0A1B2AK30 Q9VEA5 E9FYU9 A0A0P4XCV5 A0A0Q9WWF1 A0A0P5H0X3 T1DH95 A0A2M4AW40 A0A2M4C2U6 W5J580 A0A182FS24 A0A2M3ZIT3 A0A182YHX8 A0A182PRU8 A0A182MSX9 A0A182SCG4 A0A182VKK5 A0A182K7I6 A0A182VW03 A0A182UAM8 A0A182X9D5 F5HMV9 A0A182RSC9 A0A182HKC1 A0A3S3PI72 A0A1J1IHK1 G3MMV2 A0A1E1XTZ6 A0A023FFE1 A0A131XG95 A0A1E1WXK1 A0A023GEV0 G3MMV1 A0A023FUH5 H3BCJ8 A0A087TQ00 A0A224YVY4 A0A131YF29 L7M2X9 T1J3Q6 K1QN72 Q6DRG4 A0A3N0YKF7 E3TD45 E3TGE1
A0A0L7LKF0 A0A2W1BVN3 A0A194QSH8 I4DK70 A0A194PNR5 A0A2J7PJH7 K7JC40 A0A0C9R9N0 A0A1B6CJH2 A0A1B6ME97 A0A1B6FAU2 A0A0P4VY13 A0A069DVZ2 R4G820 A0A0V0G8N2 A0A023FAV0 A0A1B6IZR8 A0A1Y1L2V3 D6WIZ2 A0A2R7X5U9 A0A023EG90 A0A0K8TS37 A0A1S3DJH0 B0WK75 Q175I0 A0A1Q3FD64 W8BN20 A0A0K8UDC6 A0A0C9QL36 A0A0A1XT68 A0A1L8DGE0 A0A1I8N4B4 A0A1I8NLM1 N6TBU0 A0A0A9XSM1 U5ERW6 A0A1W4UK81 A0A0Q9WH43 A0A0Q9X0N4 A0A0R3NNP8 A0A0R1E0T3 A0A0P8XTR8 A0A336M5M8 A0A0P6DGL0 A0A0P5MMG7 A0A182NQR8 A0A182QJ50 A0A182IXN9 C3XPX8 A0A1B2AK30 Q9VEA5 E9FYU9 A0A0P4XCV5 A0A0Q9WWF1 A0A0P5H0X3 T1DH95 A0A2M4AW40 A0A2M4C2U6 W5J580 A0A182FS24 A0A2M3ZIT3 A0A182YHX8 A0A182PRU8 A0A182MSX9 A0A182SCG4 A0A182VKK5 A0A182K7I6 A0A182VW03 A0A182UAM8 A0A182X9D5 F5HMV9 A0A182RSC9 A0A182HKC1 A0A3S3PI72 A0A1J1IHK1 G3MMV2 A0A1E1XTZ6 A0A023FFE1 A0A131XG95 A0A1E1WXK1 A0A023GEV0 G3MMV1 A0A023FUH5 H3BCJ8 A0A087TQ00 A0A224YVY4 A0A131YF29 L7M2X9 T1J3Q6 K1QN72 Q6DRG4 A0A3N0YKF7 E3TD45 E3TGE1
PDB
2C35
E-value=3.73926e-38,
Score=390
Ontologies
GO
Topology
Subcellular location
Nucleus
Chromosome
Chromosome
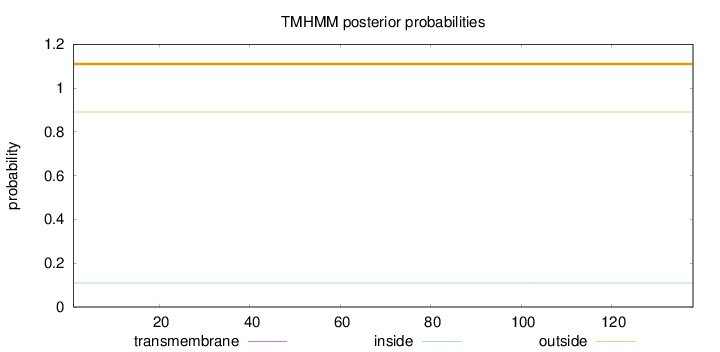
Length:
138
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10960
outside
1 - 138
Population Genetic Test Statistics
Pi
312.11101
Theta
210.355221
Tajima's D
1.501066
CLR
0.002818
CSRT
0.788610569471526
Interpretation
Uncertain
