Gene
KWMTBOMO04546 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005468
Annotation
PREDICTED:_clustered_mitochondria_protein_homolog_[Amyelois_transitella]
Full name
Clustered mitochondria protein homolog
+ More
Protein clueless
Protein clueless
Alternative Name
Clustered mitochondria protein homolog
Location in the cell
Nuclear Reliability : 2.73
Sequence
CDS
ATGAAGAAAGATGTCTGCCCGAAGATGGCATCCGCCAGCAACGGTTATGCTAACAGTCTTAACGGCCGCGCTGATGACGAGGGTGGTGGCGAGAGTGGCACGCCGCGTGTTTCCTACGCGGCTGCCGCTCGTAAGCCAGTCTCGCCTCAGTCTAGTAACACTCCTGCATCTTTTACGCTAAATACAGAACACCAGAAGTCGAAGATCAATGTTGAAAAGGAATCACAACAAAAATTAATAAATTCTACAACAAACGTCAACGGTATGGCTTCAATATCAAACGATATTGAAATAAATAATTCTAAATTAACTATCAAAGAGCCAGCAGTAAATGGCACGGAAAAAAGCACTGAGACCATAATAAATGGCGATTCAAAAAACGTTTTAGATTGTGATACTAGAAGTAGTGATAATACTAAAGTAGTTGTAAACGGCAATAATAGTGATTCCGAAGAAACTAAATCTACGAAGTCGGAAGAAGATCTTAAAACTGTTATAGTAACAAAATGTCCAAATAAAACGGCCGTGATGGACAAAAAGAAAGAACACAAGAAGAACAGGTCTTTGGAAAAGTGCAATGACGCTGAAAAGAAGACTGCTCAAAACGTAGTGGAGGATAAAAAAGACAATAGTTCATCTGTTAATATAAATAATGCCATAGAAAAAAATAATAGTGATTCAAATAAAGTGGTTAATGGCGATAGAGATAGGGAGTCCTCGCCTAGTGAAGATGGCGATGAAAAGAAAAGGGATGCTGAGGTCGTGTTTATACAAGACTTGGGTTTTACGGTTAAAATCGTTAGCCCTGGAGCTGAACCATTGGATATACAAGTATCAAGTATGGAACTTGTACAAGAAATACATCAAGTACTGATGGATCGAGAAGATACATGTCATCGAACGTGCTTTTCACTGCAACTGGATGGAGTTACTTTGGATAATTTTGCCGAATTGAAGAATATTGAAGGGCTAAAAGAAGGTTCTGTTATTAAGGTGATGGAAGAGCCGTACACAATGCGAGAAGCGCGTATCCACGTTCGACATGTTAGAGATCTTCTGAAATCCATCGATTATACTGATGCTTATGCCGGTCAAGAGTGTAGTTCATTGGCTTTTCTGAACGTCATCACGCAAGGAGATATTTTAGAAAAGAAGAAATCTCGACCAGAAAGCGTGGACTGTACTCCTCCAGACTACATAATGCCGTCCAGCACGGAGCGTCCACTGTTGCCGCTACACCCGGGACTCACTAAAGAGAATAAGGCGCCGCAGTGTCTCAAAGTACTGACCACATCGGGCTGGAATCCTCCACCTGGTCCTAGGAAGATGTGCGGAGACCTTCTTTATTTACACGTGGTGACCCTTGAAGATCGACACTTCCACATAACGGCCTGCCCTAGGGGCTTCTACCTCAACCAATCCACCGAAGAAGTGTTCAATCCTCGACCAGCTAATCCGTCTTTACTTTGTCACTCGCTCATTGAACTCCTGAGCATCGTGTCGCCGGCCTTCAAACGGAACTTCGCGCTAGTACAAAAGCGGCGTATGCAGAAACATCCGTTCGAGAGGGTCGCCACACCATACCAGGTTTATCAGTGGGCCTCGCCTATGCTCGATCACACTGTTGACGCCATCCGTGCTGAAGACACATTTTCATCGAAATTGGGCTACGAAGAGCATATTCCTGGTCAAACCCGAGACTGGAATGAGGAGTTGCAGACAACGCGCGAGTTACCAAGGTCCACTTTACCCGAGCGACTTCTGCGTGAAAGAGCCATTTTTAAGGTCCACAGTGATTTCGTTGCTGCCGCGACTCGAGGAGCCATGGCTGTGGTCGATGGTAACGTGATGGCGATCAATCCAGGGGAAGAACCGAAGATGCAGATGTTCATCTGGAACAATATCTTCTTCTCGTTGGGCTTCGACGTACGCGATCACTACAAAGATTTGGGCGGAGATGCGGCGGCTTTTGTTGCTCCGCGCAATGACTTGCAAGGCGTTCGAGTGTACAGCGCAGTAGACACCCCGGGGCTGCACACCCTGGGCACGGTGGTGGTGGACTACCGCGGGTACCGGGTCACGGCTCAGTCCATCATCCCGGGAATACTGGAGAAGGAGCAAGAACAGAGCGTTGTATACGGCAGCATCGACTTTGGAACGACCGTACTCTCGCACCCTAAGTACATGGAACTGTTGAGCAAAGCTGGACAGCAGTTAAAGATAATGCCCCATTCAGTTATCAGTGCGAATGGCGAGGTGGTTGAGTTGTGCTCCAGCGTTGAATGCAAGGGAATTATAGGAAACGACGGACGTCATTACATTTTAGACTTGCTGCGCACTTTTCCACCGGATGTCAACTTTTTACAATTGGAAGATGACGAACTCAGAGAAGATATAAAATCCATGGGCTTCCCGATTATCCACAAGCACAAACTATGTTGTTTGAGACAGGAATTAGTCGATAGTTTTGTTGAGGCTCGTTATTTCATGTTCATTCGCTATGCCGCGTTCCACTTGCAACAGTTAAGCGCGAAGCGTCAGCGCGATCAGAATGATGATCAGAAGTCAATCGAACCGAACAAGGAAAGCGAAAAGAAAGATAATACAGACTCAGAGAAGAAGAAGAGAGACAATAAAAGCCAAGAGAAAGAGACCAAAGATAAAAAGAAAAAGGATGAATCTAGTAGTGTTAAAAAAGAAGAAAGTAAGAAAGAAATTGAATTAAATGATGTTAAGATTGAGAAAGGGAAAGATGATGAATTATTGTTGAACGACAACTATTCAGAAATAGACACGGATGTTGCCAAGAAAATTGTTGAAAGCATTACGGACTCAATTTGCAGCGGAGAGAAACAAGAGAATGACTCGGGCGAGCGTTCCCGCGCAGTGGTGGCGGCGGCGGCGCGCGCTGTGGCCTCGCTCAAGGAGTCGGAGTTCGACGTGCGCTTCAACCCGGACGTGTACTCCGCCGGCATCAAGCACGCCGCCGCGCCCGAGCACCTCGCCAAGCAACGGCACCTCGTCAAGGAAGCGGCCGCCTTCCTGCTCACCACGCAGATCCCGGCCTTCGTGCGGGAGTGCGTGGAGCAGAGCGTGGCGCCGATGGACGGCGCGGGGCTGACGGAGGCGCTGCACGCGCGCGGCATCAACGTGCGGTACCTGGGCCGCGTGGCGCTGGCCCTGGCTGCGCACCCCTCGCTGGCCTACCTGCACGCCATCGCGCTCGCCGACCTGCTGCTGCGCGCCGCCAAGCATCTCTACACGGCCTACTTGCAGGGTTGCGAGCCTATGTGCATCGGTTCAGCTGTAGCGCATTTCTTGAACTGTTTGTTGGGGGCGTGTCCGACGCCCGGCATCAGTCCGGCGGAGGGCGTGGCGGGGGGTCGGGGCGCGGGGCGCGGGGGTCGCGGGCGCCGCTCCCGCAAGCACGCGCACTCGCAGCCCGCCGCCTCGCCGCTCCCCGACTGGCAGAACCTCACGCCCAAATCGCTCTTTGCACAGATTAAACAAGAGTTGAAGGCGTATTGGGGATATGATCTTAATGCCGAGAACATGGAATCTATAATCGAAAAACACAGTATTCAGAAAATATCACTTCTCAGGTCCTTCGCACTAAAAGTAGGCTTGCAAGTTATGTTAAGAGAATACGACTTTGACAATAAAAACAAGGCACCGTTTGCAAGTGCGGACATTATGAATATTTTCCCCGTCGTTAAACATATCAATCCACGCGCTTCAGACGCATACAATTTCTACACGACGGGTCAGAACAAAATCCAAGCCGGCGCCGTGTCCGAAGGTCACGAACTGATCGCTGAGGCTCTGAACTTGCTGAACAACGTGTACGGCGCCATGCACGGCGAGATCGCGCAGTGCCTCCGCATGGTGGCGCGCCTGTGCTACGTCACCGGCGAGCACCGGGACGCCATGGCCTACCAGCAGAAGGCGGTGCTTATGTCCGAGAGGGTCAACGGCATTGATCACCCTTACACTATTACAGAATATTCACATTTGGCGTTATACTGCTTTGCCAACGGTCAAGTGAGCACCGCATTGAAGCTACTGTATCGAGCCCGCTACTTGGCATTGATCGTTTGCGGCGAAAACCACCCAGAGATGGCGCTGCTCGACAGCAACATCGCGCTGATCCTGCACGCGGTCGGCGAGTACGAGCTGTCGCTGCGGTTCGCGGAGCGCGGGCTGGCGGTGACGAGCGGCACGCACGGGCCGCGCTCGCTGAAGGCGGCCGTGGCGCGCCACCTGCTCGCGCGCACGCTCTCCTGCCTCGGCGACTTCCGGGCGGCGCTCGCGCACGAGAAGGAGACCTACTCTATCTATAAGCATCTGTTGGGCGAGAAGCACGAGAAGACGCGCGAGTCGTCGGAGTGTCTCCGTCACCTGACGCAGCAGGCGGTGGTGCTGCAGAAGCGGCTGGCGGAGGCGTACGCGCGCGCCCCGCACCACGCGCACCACGCGCCGCTGCACATCCAGCCGCCCGGCATGGCCTCCGTCATCGACATGCTCAACCTCATCAACGGTATACTCTTCGTGCAAATCAGCCCGCAAGACATCGAGCAGTTCAAAGCTGAGATAGAGAAAAGACAACTGAAGGATTTGCCCGTTCCTGGAACTCTGACCGATCTCGGCGGAGAAGTCGACGACGAGAAATCTAATTCCGGCGCGCCGTCCGCGAGCGGCGACACGAAGGAGGCGCCGGCCAGTTGA
Protein
MKKDVCPKMASASNGYANSLNGRADDEGGGESGTPRVSYAAAARKPVSPQSSNTPASFTLNTEHQKSKINVEKESQQKLINSTTNVNGMASISNDIEINNSKLTIKEPAVNGTEKSTETIINGDSKNVLDCDTRSSDNTKVVVNGNNSDSEETKSTKSEEDLKTVIVTKCPNKTAVMDKKKEHKKNRSLEKCNDAEKKTAQNVVEDKKDNSSSVNINNAIEKNNSDSNKVVNGDRDRESSPSEDGDEKKRDAEVVFIQDLGFTVKIVSPGAEPLDIQVSSMELVQEIHQVLMDREDTCHRTCFSLQLDGVTLDNFAELKNIEGLKEGSVIKVMEEPYTMREARIHVRHVRDLLKSIDYTDAYAGQECSSLAFLNVITQGDILEKKKSRPESVDCTPPDYIMPSSTERPLLPLHPGLTKENKAPQCLKVLTTSGWNPPPGPRKMCGDLLYLHVVTLEDRHFHITACPRGFYLNQSTEEVFNPRPANPSLLCHSLIELLSIVSPAFKRNFALVQKRRMQKHPFERVATPYQVYQWASPMLDHTVDAIRAEDTFSSKLGYEEHIPGQTRDWNEELQTTRELPRSTLPERLLRERAIFKVHSDFVAAATRGAMAVVDGNVMAINPGEEPKMQMFIWNNIFFSLGFDVRDHYKDLGGDAAAFVAPRNDLQGVRVYSAVDTPGLHTLGTVVVDYRGYRVTAQSIIPGILEKEQEQSVVYGSIDFGTTVLSHPKYMELLSKAGQQLKIMPHSVISANGEVVELCSSVECKGIIGNDGRHYILDLLRTFPPDVNFLQLEDDELREDIKSMGFPIIHKHKLCCLRQELVDSFVEARYFMFIRYAAFHLQQLSAKRQRDQNDDQKSIEPNKESEKKDNTDSEKKKRDNKSQEKETKDKKKKDESSSVKKEESKKEIELNDVKIEKGKDDELLLNDNYSEIDTDVAKKIVESITDSICSGEKQENDSGERSRAVVAAAARAVASLKESEFDVRFNPDVYSAGIKHAAAPEHLAKQRHLVKEAAAFLLTTQIPAFVRECVEQSVAPMDGAGLTEALHARGINVRYLGRVALALAAHPSLAYLHAIALADLLLRAAKHLYTAYLQGCEPMCIGSAVAHFLNCLLGACPTPGISPAEGVAGGRGAGRGGRGRRSRKHAHSQPAASPLPDWQNLTPKSLFAQIKQELKAYWGYDLNAENMESIIEKHSIQKISLLRSFALKVGLQVMLREYDFDNKNKAPFASADIMNIFPVVKHINPRASDAYNFYTTGQNKIQAGAVSEGHELIAEALNLLNNVYGAMHGEIAQCLRMVARLCYVTGEHRDAMAYQQKAVLMSERVNGIDHPYTITEYSHLALYCFANGQVSTALKLLYRARYLALIVCGENHPEMALLDSNIALILHAVGEYELSLRFAERGLAVTSGTHGPRSLKAAVARHLLARTLSCLGDFRAALAHEKETYSIYKHLLGEKHEKTRESSECLRHLTQQAVVLQKRLAEAYARAPHHAHHAPLHIQPPGMASVIDMLNLINGILFVQISPQDIEQFKAEIEKRQLKDLPVPGTLTDLGGEVDDEKSNSGAPSASGDTKEAPAS
Summary
Description
mRNA-binding protein involved in proper cytoplasmic distribution of mitochondria.
Similarity
Belongs to the CLU family.
Keywords
Complete proteome
Cytoplasm
Reference proteome
Repeat
TPR repeat
Phosphoprotein
Feature
chain Clustered mitochondria protein homolog
Uniprot
H9J7H6
A0A2W1BYT0
A0A3S2M5A8
A0A212EQM5
A0A067QPB0
A0A1B6BYB3
+ More
A0A1B6E4P9 A0A1B6LQ96 A0A1B6KN93 A0A1B6KI19 D6WEY1 E2AE38 E9IWH2 A0A3L8DAF3 A0A0J7KSX0 T1HWY7 E2BKC5 A0A0V0G6S6 A0A224XGI9 A0A0L7R8M6 A0A023F4P9 A0A158NF15 A0A088APW1 A0A2A3EIK1 A0A232FHW9 A0A1W4WYU9 A0A0P4VY90 A0A026W381 E0VTU9 A0A1Y1N549 A0A0N0BC92 A0A0N8DGS0 A0A0P5NTI8 E9GHX8 A0A162T1D9 A0A1J1II94 A0A1W4WMT7 A0A0P5PIN1 A0A0P5Z701 J9JPN1 A0A2H8TIW5 Q17N71 A0A0N8DE08 A0A087U7G8 A0A182G693 B0W2S0 A0A0P6GK75 A0A336MJK7 A0A0P5QNL8 A0A336MCW9 A0A0P4ZVD9 A0A1Q3F5P5 A0A1Q3F5Z6 A0A1Q3F5Z8 A0A0P5HA82 U4U3P3 N6TNQ3 A0A0P5A0J9 A0A151XIF5 A0A1L8DS54 T1ITX2 A0A0P6HVZ1 A0A2M4CZ13 W4VRL3 A0A182JEF2 A0A2M4A682 A0A2M4A6C8 A0A2M4B947 W5JQY0 A0A0K2T2F5 A0A182MC92 A0A0P5HMQ9 A0A2M4CYZ7 A0A0P5J1J4 T1PBX9 A0A0P5H9J3 A0A182R4P3 A0A182JQA8 A0A182P4Y9 A0A182Y9D1 A0A0P5HEJ2 A0A1I8MCE8 Q7PZD5 A0A182UKA1 A0A182US05 A0A182QP10 A0A084WIS2 A0A182N3Y6 A0A1B0C267 A0A182HHG4 A0A182XGU6 A0A1A9YLF6 T1JVZ2 A0A131XMR2 A0A131YIU2 Q291J5 B4GAM1 A0A3B0JH73 A0A0R3NLP5 A0A182L4Z1
A0A1B6E4P9 A0A1B6LQ96 A0A1B6KN93 A0A1B6KI19 D6WEY1 E2AE38 E9IWH2 A0A3L8DAF3 A0A0J7KSX0 T1HWY7 E2BKC5 A0A0V0G6S6 A0A224XGI9 A0A0L7R8M6 A0A023F4P9 A0A158NF15 A0A088APW1 A0A2A3EIK1 A0A232FHW9 A0A1W4WYU9 A0A0P4VY90 A0A026W381 E0VTU9 A0A1Y1N549 A0A0N0BC92 A0A0N8DGS0 A0A0P5NTI8 E9GHX8 A0A162T1D9 A0A1J1II94 A0A1W4WMT7 A0A0P5PIN1 A0A0P5Z701 J9JPN1 A0A2H8TIW5 Q17N71 A0A0N8DE08 A0A087U7G8 A0A182G693 B0W2S0 A0A0P6GK75 A0A336MJK7 A0A0P5QNL8 A0A336MCW9 A0A0P4ZVD9 A0A1Q3F5P5 A0A1Q3F5Z6 A0A1Q3F5Z8 A0A0P5HA82 U4U3P3 N6TNQ3 A0A0P5A0J9 A0A151XIF5 A0A1L8DS54 T1ITX2 A0A0P6HVZ1 A0A2M4CZ13 W4VRL3 A0A182JEF2 A0A2M4A682 A0A2M4A6C8 A0A2M4B947 W5JQY0 A0A0K2T2F5 A0A182MC92 A0A0P5HMQ9 A0A2M4CYZ7 A0A0P5J1J4 T1PBX9 A0A0P5H9J3 A0A182R4P3 A0A182JQA8 A0A182P4Y9 A0A182Y9D1 A0A0P5HEJ2 A0A1I8MCE8 Q7PZD5 A0A182UKA1 A0A182US05 A0A182QP10 A0A084WIS2 A0A182N3Y6 A0A1B0C267 A0A182HHG4 A0A182XGU6 A0A1A9YLF6 T1JVZ2 A0A131XMR2 A0A131YIU2 Q291J5 B4GAM1 A0A3B0JH73 A0A0R3NLP5 A0A182L4Z1
Pubmed
EMBL
BABH01013845
BABH01013846
BABH01013847
BABH01013848
KZ149904
PZC78397.1
+ More
RSAL01000036 RVE51305.1 AGBW02013261 OWR43803.1 KK853109 KDR11240.1 GEDC01031026 JAS06272.1 GEDC01004439 JAS32859.1 GEBQ01014076 JAT25901.1 GEBQ01027081 JAT12896.1 GEBQ01028876 JAT11101.1 KQ971318 EFA00454.1 GL438827 EFN68271.1 GL766526 EFZ15097.1 QOIP01000010 RLU17475.1 LBMM01003605 KMQ93329.1 ACPB03000369 ACPB03000370 ACPB03000371 ACPB03000372 GL448783 EFN83849.1 GECL01002414 JAP03710.1 GFTR01008724 JAW07702.1 KQ414632 KOC67121.1 GBBI01002325 JAC16387.1 ADTU01013696 ADTU01013697 KZ288232 PBC31528.1 NNAY01000187 OXU30173.1 GDKW01002045 JAI54550.1 KK107455 EZA50535.1 DS235773 EEB16805.1 GEZM01012263 JAV93031.1 KQ435922 KOX68642.1 GDIP01035287 JAM68428.1 GDIQ01137966 JAL13760.1 GL732545 EFX80971.1 LRGB01000024 KZS21814.1 CVRI01000047 CRK98177.1 GDIQ01149420 JAL02306.1 GDIP01048234 JAM55481.1 ABLF02034686 GFXV01002249 MBW14054.1 CH477201 GDIP01042983 JAM60732.1 KK118574 KFM73307.1 JXUM01147461 KQ570372 KXJ68378.1 DS231828 GDIQ01033510 JAN61227.1 UFQT01000661 SSX26248.1 GDIQ01111764 JAL39962.1 SSX26247.1 GDIP01207515 JAJ15887.1 GFDL01012168 JAV22877.1 GFDL01012068 JAV22977.1 GFDL01012059 JAV22986.1 GDIQ01232882 JAK18843.1 KB631604 ERL84600.1 APGK01057365 KB741280 ENN70880.1 GDIP01210061 JAJ13341.1 KQ982080 KYQ60186.1 GFDF01004815 JAV09269.1 JH431506 GDIQ01021315 JAN73422.1 GGFL01006327 MBW70505.1 GANO01001749 JAB58122.1 GGFK01002993 MBW36314.1 GGFK01002951 MBW36272.1 GGFJ01000416 MBW49557.1 ADMH02000368 ETN66807.1 HACA01002852 CDW20213.1 AXCM01006955 GDIQ01230927 JAK20798.1 GGFL01006317 MBW70495.1 GDIQ01230928 JAK20797.1 KA646272 AFP60901.1 GDIQ01230929 JAK20796.1 GDIQ01230926 JAK20799.1 AAAB01008986 AXCN02001095 ATLV01023945 KE525347 KFB50116.1 JXJN01021060 JXJN01024363 APCN01002408 CAEY01000798 GEFH01000969 JAP67612.1 GEDV01010187 JAP78370.1 CM000071 CH479181 OUUW01000001 SPP74730.1 KRT01854.1
RSAL01000036 RVE51305.1 AGBW02013261 OWR43803.1 KK853109 KDR11240.1 GEDC01031026 JAS06272.1 GEDC01004439 JAS32859.1 GEBQ01014076 JAT25901.1 GEBQ01027081 JAT12896.1 GEBQ01028876 JAT11101.1 KQ971318 EFA00454.1 GL438827 EFN68271.1 GL766526 EFZ15097.1 QOIP01000010 RLU17475.1 LBMM01003605 KMQ93329.1 ACPB03000369 ACPB03000370 ACPB03000371 ACPB03000372 GL448783 EFN83849.1 GECL01002414 JAP03710.1 GFTR01008724 JAW07702.1 KQ414632 KOC67121.1 GBBI01002325 JAC16387.1 ADTU01013696 ADTU01013697 KZ288232 PBC31528.1 NNAY01000187 OXU30173.1 GDKW01002045 JAI54550.1 KK107455 EZA50535.1 DS235773 EEB16805.1 GEZM01012263 JAV93031.1 KQ435922 KOX68642.1 GDIP01035287 JAM68428.1 GDIQ01137966 JAL13760.1 GL732545 EFX80971.1 LRGB01000024 KZS21814.1 CVRI01000047 CRK98177.1 GDIQ01149420 JAL02306.1 GDIP01048234 JAM55481.1 ABLF02034686 GFXV01002249 MBW14054.1 CH477201 GDIP01042983 JAM60732.1 KK118574 KFM73307.1 JXUM01147461 KQ570372 KXJ68378.1 DS231828 GDIQ01033510 JAN61227.1 UFQT01000661 SSX26248.1 GDIQ01111764 JAL39962.1 SSX26247.1 GDIP01207515 JAJ15887.1 GFDL01012168 JAV22877.1 GFDL01012068 JAV22977.1 GFDL01012059 JAV22986.1 GDIQ01232882 JAK18843.1 KB631604 ERL84600.1 APGK01057365 KB741280 ENN70880.1 GDIP01210061 JAJ13341.1 KQ982080 KYQ60186.1 GFDF01004815 JAV09269.1 JH431506 GDIQ01021315 JAN73422.1 GGFL01006327 MBW70505.1 GANO01001749 JAB58122.1 GGFK01002993 MBW36314.1 GGFK01002951 MBW36272.1 GGFJ01000416 MBW49557.1 ADMH02000368 ETN66807.1 HACA01002852 CDW20213.1 AXCM01006955 GDIQ01230927 JAK20798.1 GGFL01006317 MBW70495.1 GDIQ01230928 JAK20797.1 KA646272 AFP60901.1 GDIQ01230929 JAK20796.1 GDIQ01230926 JAK20799.1 AAAB01008986 AXCN02001095 ATLV01023945 KE525347 KFB50116.1 JXJN01021060 JXJN01024363 APCN01002408 CAEY01000798 GEFH01000969 JAP67612.1 GEDV01010187 JAP78370.1 CM000071 CH479181 OUUW01000001 SPP74730.1 KRT01854.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000027135
UP000007266
UP000000311
+ More
UP000279307 UP000036403 UP000015103 UP000008237 UP000053825 UP000005205 UP000005203 UP000242457 UP000215335 UP000192223 UP000053097 UP000009046 UP000053105 UP000000305 UP000076858 UP000183832 UP000007819 UP000008820 UP000054359 UP000069940 UP000249989 UP000002320 UP000030742 UP000019118 UP000075809 UP000075880 UP000000673 UP000075883 UP000075900 UP000075881 UP000075885 UP000076408 UP000095301 UP000007062 UP000075902 UP000075903 UP000075886 UP000030765 UP000075884 UP000092460 UP000075840 UP000076407 UP000092443 UP000015104 UP000001819 UP000008744 UP000268350 UP000075882
UP000279307 UP000036403 UP000015103 UP000008237 UP000053825 UP000005205 UP000005203 UP000242457 UP000215335 UP000192223 UP000053097 UP000009046 UP000053105 UP000000305 UP000076858 UP000183832 UP000007819 UP000008820 UP000054359 UP000069940 UP000249989 UP000002320 UP000030742 UP000019118 UP000075809 UP000075880 UP000000673 UP000075883 UP000075900 UP000075881 UP000075885 UP000076408 UP000095301 UP000007062 UP000075902 UP000075903 UP000075886 UP000030765 UP000075884 UP000092460 UP000075840 UP000076407 UP000092443 UP000015104 UP000001819 UP000008744 UP000268350 UP000075882
PRIDE
Pfam
Interpro
IPR027523
CLU
+ More
IPR011990 TPR-like_helical_dom_sf
IPR028275 CLU_N
IPR033646 CLU-central
IPR025697 CLU_dom
IPR023231 GSKIP_dom_sf
IPR007967 GSKIP_dom
IPR013026 TPR-contain_dom
IPR031311 CHIT_BIND_RR_consensus
IPR000618 Insect_cuticle
IPR012926 TMPIT
IPR019734 TPR_repeat
IPR012934 Znf_AD
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR036959 Peptidase_C12_UCH_sf
IPR001578 Peptidase_C12_UCH
IPR038765 Papain-like_cys_pep_sf
IPR041507 UCH_C
IPR011990 TPR-like_helical_dom_sf
IPR028275 CLU_N
IPR033646 CLU-central
IPR025697 CLU_dom
IPR023231 GSKIP_dom_sf
IPR007967 GSKIP_dom
IPR013026 TPR-contain_dom
IPR031311 CHIT_BIND_RR_consensus
IPR000618 Insect_cuticle
IPR012926 TMPIT
IPR019734 TPR_repeat
IPR012934 Znf_AD
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR036959 Peptidase_C12_UCH_sf
IPR001578 Peptidase_C12_UCH
IPR038765 Papain-like_cys_pep_sf
IPR041507 UCH_C
Gene 3D
ProteinModelPortal
H9J7H6
A0A2W1BYT0
A0A3S2M5A8
A0A212EQM5
A0A067QPB0
A0A1B6BYB3
+ More
A0A1B6E4P9 A0A1B6LQ96 A0A1B6KN93 A0A1B6KI19 D6WEY1 E2AE38 E9IWH2 A0A3L8DAF3 A0A0J7KSX0 T1HWY7 E2BKC5 A0A0V0G6S6 A0A224XGI9 A0A0L7R8M6 A0A023F4P9 A0A158NF15 A0A088APW1 A0A2A3EIK1 A0A232FHW9 A0A1W4WYU9 A0A0P4VY90 A0A026W381 E0VTU9 A0A1Y1N549 A0A0N0BC92 A0A0N8DGS0 A0A0P5NTI8 E9GHX8 A0A162T1D9 A0A1J1II94 A0A1W4WMT7 A0A0P5PIN1 A0A0P5Z701 J9JPN1 A0A2H8TIW5 Q17N71 A0A0N8DE08 A0A087U7G8 A0A182G693 B0W2S0 A0A0P6GK75 A0A336MJK7 A0A0P5QNL8 A0A336MCW9 A0A0P4ZVD9 A0A1Q3F5P5 A0A1Q3F5Z6 A0A1Q3F5Z8 A0A0P5HA82 U4U3P3 N6TNQ3 A0A0P5A0J9 A0A151XIF5 A0A1L8DS54 T1ITX2 A0A0P6HVZ1 A0A2M4CZ13 W4VRL3 A0A182JEF2 A0A2M4A682 A0A2M4A6C8 A0A2M4B947 W5JQY0 A0A0K2T2F5 A0A182MC92 A0A0P5HMQ9 A0A2M4CYZ7 A0A0P5J1J4 T1PBX9 A0A0P5H9J3 A0A182R4P3 A0A182JQA8 A0A182P4Y9 A0A182Y9D1 A0A0P5HEJ2 A0A1I8MCE8 Q7PZD5 A0A182UKA1 A0A182US05 A0A182QP10 A0A084WIS2 A0A182N3Y6 A0A1B0C267 A0A182HHG4 A0A182XGU6 A0A1A9YLF6 T1JVZ2 A0A131XMR2 A0A131YIU2 Q291J5 B4GAM1 A0A3B0JH73 A0A0R3NLP5 A0A182L4Z1
A0A1B6E4P9 A0A1B6LQ96 A0A1B6KN93 A0A1B6KI19 D6WEY1 E2AE38 E9IWH2 A0A3L8DAF3 A0A0J7KSX0 T1HWY7 E2BKC5 A0A0V0G6S6 A0A224XGI9 A0A0L7R8M6 A0A023F4P9 A0A158NF15 A0A088APW1 A0A2A3EIK1 A0A232FHW9 A0A1W4WYU9 A0A0P4VY90 A0A026W381 E0VTU9 A0A1Y1N549 A0A0N0BC92 A0A0N8DGS0 A0A0P5NTI8 E9GHX8 A0A162T1D9 A0A1J1II94 A0A1W4WMT7 A0A0P5PIN1 A0A0P5Z701 J9JPN1 A0A2H8TIW5 Q17N71 A0A0N8DE08 A0A087U7G8 A0A182G693 B0W2S0 A0A0P6GK75 A0A336MJK7 A0A0P5QNL8 A0A336MCW9 A0A0P4ZVD9 A0A1Q3F5P5 A0A1Q3F5Z6 A0A1Q3F5Z8 A0A0P5HA82 U4U3P3 N6TNQ3 A0A0P5A0J9 A0A151XIF5 A0A1L8DS54 T1ITX2 A0A0P6HVZ1 A0A2M4CZ13 W4VRL3 A0A182JEF2 A0A2M4A682 A0A2M4A6C8 A0A2M4B947 W5JQY0 A0A0K2T2F5 A0A182MC92 A0A0P5HMQ9 A0A2M4CYZ7 A0A0P5J1J4 T1PBX9 A0A0P5H9J3 A0A182R4P3 A0A182JQA8 A0A182P4Y9 A0A182Y9D1 A0A0P5HEJ2 A0A1I8MCE8 Q7PZD5 A0A182UKA1 A0A182US05 A0A182QP10 A0A084WIS2 A0A182N3Y6 A0A1B0C267 A0A182HHG4 A0A182XGU6 A0A1A9YLF6 T1JVZ2 A0A131XMR2 A0A131YIU2 Q291J5 B4GAM1 A0A3B0JH73 A0A0R3NLP5 A0A182L4Z1
PDB
6FV0
E-value=9.71288e-07,
Score=132
Ontologies
GO
GO:0005737
GO:0048312
GO:0003723
GO:0007005
GO:0003743
GO:0003729
GO:0042302
GO:0016021
GO:0005634
GO:0008270
GO:0004843
GO:0006511
GO:0043022
GO:0005794
GO:0055059
GO:0005783
GO:0098799
GO:0072686
GO:0031315
GO:0005515
GO:0030131
GO:0030117
GO:0005488
GO:0008565
GO:0015031
GO:0016020
GO:0016742
GO:0003824
Topology
Subcellular location
Cytoplasm
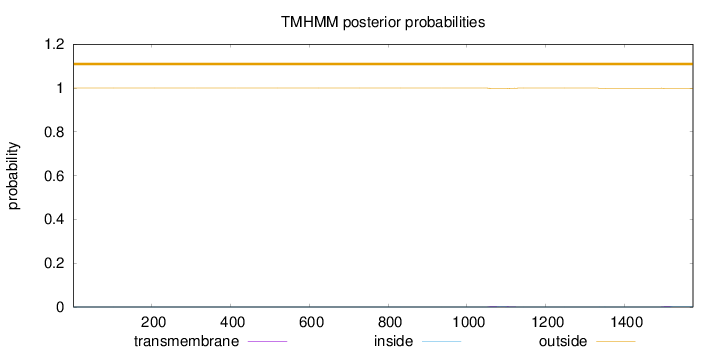
Length:
1574
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11136
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00003
outside
1 - 1574
Population Genetic Test Statistics
Pi
241.187358
Theta
175.270601
Tajima's D
1.027187
CLR
0.344062
CSRT
0.671116444177791
Interpretation
Uncertain
