Gene
KWMTBOMO04543 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005466
Annotation
PREDICTED:_AP-2_complex_subunit_alpha_[Papilio_xuthus]
Full name
AP-2 complex subunit alpha
Alternative Name
Alpha-adaptin
Location in the cell
Cytoplasmic Reliability : 1.232 Golgi Reliability : 1.838
Sequence
CDS
ATGCCTGCCGTCAGAGGAGATGGTATGCGAGGGCTCGCTGTCTTTATCTCAGACATAAGAAACTGTAAGAGTAAAGAAGCAGAAATAAAGAGAATCAACAAGGAGCTTGCAAATATAAGAAGTAAGTTTAAGGGTGATAAAACTCTAGATGGATATCAAAAGAAGAAGTATGTGTGTAAACTTCTGTTTATATTTCTGCTCGGACATGATATCGACTTCGGGCACATGGAAGCAGTAAACTTACTTTCATCCAATAAATACTCCGAAAAACAGATTGGATACCTTTTCATATCTGTCCTAGTCAATACCAACAGTGACCTCATTAAACTTATCATTCAGAGCATTAAAAACGACTTGCAGTCTCGCAATCCTATTCATGTAAACTTAGCTCTTCAATGTATTGCCAACATTGGTAGCAAAGATATGGCTGAAGCTTTTGGTACTGAAATACCCAAACTTCTTGTCTCTGGTGATACCATGGATGTAGTGAAACAATCTGCTGCACTTTGCCTCCTTCGCTTATTTCGCAAATCTCCTGAGATAATACCTGGAGGAGAGTGGACTTCTCGCATAATTCATCTTTTGAATGACCCCCATATGGGTGTAGTAACTGCAGCGACATCTTTGATTGATGCTCTTGTAAAGAAGAATCCTGATGAGTATAAGGGTTGTGTAACATTGGCTGTAGCTCGACTTAGCAGAATTGTTACTGCCAGTTACACAGATTTACAAGATTATACATATTACTTTGTACCTGCTCCTTGGTTGTCTGTCAAACTGTTACGCCTCTTACAAAACTACACCCCTCCATCTGAAGAACCTGGAGTTCGAGGACGTCTTTCAGAATGCCTAGAAACAATATTCAACAAAGCTCAAGAACCACCAAAATCTAAAAAAGTGCAACATTCCAATGCCAAGAATGCTGTTCTTTTTGAAGCAATAAGTCTTATTATACACAACGACAGTGAACCTAACTTATTAGTTCGTGCTTGCAATCAGCTGGGACAATTTCTTAGTAATCGTGAAACTAACTTGCGCTATTTAGCTCTTGAATCAATGTGTCATTTAGCTACATCAGAATTTTCTCATGAAGCAGTAAAAAAGCATCAAGAAATAGTAATCCTCTCAATGAAAATGGAAAAAGATGTTTCTGTCAGGCAGCAGGCTGTAGATCTTCTGTATGCAATGTGTGATAAGACTAATGCAGAAGAAATTGTACAAGAAATGTTAGCTTATTTAGAAACTGCTGATTATTCAATCAGAGAAGAAATGGTGTTAAAAGTTGCTATATTAGCAGAAAAGTATGCTACAGATTTCACTTGGTATGTGGATGTAATTCTTAATCTTATAAGAATTGCTGGTGATTATGTCTCAGAAGAAGTTTGGTACAGAGTAATTCAAATAGTAATAAATAGAGATGAAGTTCAAGCATATGCTGCTAAAACTGTATTTGAAGCATTACAAGCCCCAACATGCCATGAGAATATGGTTAAAGTTGGAGGATACATTCTGGGAGAGTTTGGTAACTTGATAGCTGGAGATACAAGATCATCACCACAAGTTCAGTTTGAACTTCTACATTCCAAATACCATTTGTGTTCTGTTGCAACAAGAGCCCTATTGTTATCAACGTATATAAAGTTGGTTAATCTTTTCCCAGAAATAAAAAATAGAGTTCAAGAAGTATTTAAGGCAGATTCTAATTTACGTTCAGCTGATGTTGAGTTGCAACAAAGGGCATCAGAATACTTGCAACTTAGTATTGTAGCCAGTACAGATGTTTTAGCCACAGTATTAGAAGAAATGCCTGCTTTCCCTGAAAGAGAATCTTCTATCTTAGCAGTATTGAAAAAGAAGAAGCCAGGTCGTATTCCAGATGATGTTAGAGAATCAAAGAGCCCTCTGCCCACTGTTGCTCCCACTGCTATACTAAATTCTACTAACAATACTAATAGCTCCAGTGCTGATCTCCTAGGACTGTCTACACCACCTGGTTCTGCAGCCCCTACTACAGGGAATGGCTTATTGGATGTACTTGGAGATTTGTACACATCAACAAAAATAAGCCCCAGTACTGTACAGCAAAATAACATCAAAAAGTTTTTGTTTAAAAACAATGGGGTGCTTTTTGAAAATGAATTGATCCAAATTGGTGTTAAAAGTGAGTTCAGACAAAATCTTGGCAGAATTGGCCTATTTTATGGCAACAAAACCCAATTTCCAATCCAGAATGTCCATCCTGAGTTACATTGGACTGACTTGCATAAGTTGAATGTACAAATGAAACCAATGGAACCTATATTGGAAGCAGGTGCTCAAATCCAACAAATGTTGACAGCTGAATGTATAGAAGATTTTACTGATGTACCAAGCATGTCAGTCTCATTTATATATAACAATGTACCACAGAAAATAACAATGAAACTCCCACTCACCCTTAACAAATTCTTTGAACCAACTGAAATGAATGGTGAATCATTCTTTGCACGGTGGAAAAATTTGGGAGGCGAACAACAGAGAGCCCAAAAGATTTTCAAAGCTCAGGGACCGATTGATTTGACTGGCACAAGAACAAAATTGGCTGGATTTGGCATGCAATTATTAGATGGTATCGATCCTAATCCAGATAACTTTGTATGTGCCGGCATTGTCCATACAAAGATGCAACAAGTTGGATGTTTAATGAGACTGGAACCTAACAAACAAGCTCAAATGTACAGACTAACCATACGATCTAGTAAGGAGTCAGTTTCTCAAGAAATCTGCAACTTACTTGCTGACCAGTTTTGA
Protein
MPAVRGDGMRGLAVFISDIRNCKSKEAEIKRINKELANIRSKFKGDKTLDGYQKKKYVCKLLFIFLLGHDIDFGHMEAVNLLSSNKYSEKQIGYLFISVLVNTNSDLIKLIIQSIKNDLQSRNPIHVNLALQCIANIGSKDMAEAFGTEIPKLLVSGDTMDVVKQSAALCLLRLFRKSPEIIPGGEWTSRIIHLLNDPHMGVVTAATSLIDALVKKNPDEYKGCVTLAVARLSRIVTASYTDLQDYTYYFVPAPWLSVKLLRLLQNYTPPSEEPGVRGRLSECLETIFNKAQEPPKSKKVQHSNAKNAVLFEAISLIIHNDSEPNLLVRACNQLGQFLSNRETNLRYLALESMCHLATSEFSHEAVKKHQEIVILSMKMEKDVSVRQQAVDLLYAMCDKTNAEEIVQEMLAYLETADYSIREEMVLKVAILAEKYATDFTWYVDVILNLIRIAGDYVSEEVWYRVIQIVINRDEVQAYAAKTVFEALQAPTCHENMVKVGGYILGEFGNLIAGDTRSSPQVQFELLHSKYHLCSVATRALLLSTYIKLVNLFPEIKNRVQEVFKADSNLRSADVELQQRASEYLQLSIVASTDVLATVLEEMPAFPERESSILAVLKKKKPGRIPDDVRESKSPLPTVAPTAILNSTNNTNSSSADLLGLSTPPGSAAPTTGNGLLDVLGDLYTSTKISPSTVQQNNIKKFLFKNNGVLFENELIQIGVKSEFRQNLGRIGLFYGNKTQFPIQNVHPELHWTDLHKLNVQMKPMEPILEAGAQIQQMLTAECIEDFTDVPSMSVSFIYNNVPQKITMKLPLTLNKFFEPTEMNGESFFARWKNLGGEQQRAQKIFKAQGPIDLTGTRTKLAGFGMQLLDGIDPNPDNFVCAGIVHTKMQQVGCLMRLEPNKQAQMYRLTIRSSKESVSQEICNLLADQF
Summary
Description
Adaptins are components of the adapter complexes which link clathrin to receptors in coated vesicles.
Adaptins are components of the adapter complexes which link clathrin to receptors in coated vesicles. Clathrin-associated protein complexes are believed to interact with the cytoplasmic tails of membrane proteins, leading to their selection and concentration. Alpha adaptin is a subunit of the plasma membrane adapter (By similarity).
Adaptins are components of the adapter complexes which link clathrin to receptors in coated vesicles. Clathrin-associated protein complexes are believed to interact with the cytoplasmic tails of membrane proteins, leading to their selection and concentration. AP-2alpha is a subunit of the plasma membrane adapter.
Adaptins are components of the adaptor complexes which link clathrin to receptors in coated vesicles. Clathrin-associated protein complexes are believed to interact with the cytoplasmic tails of membrane proteins, leading to their selection and concentration.
Adaptins are components of the adapter complexes which link clathrin to receptors in coated vesicles. Clathrin-associated protein complexes are believed to interact with the cytoplasmic tails of membrane proteins, leading to their selection and concentration. Alpha adaptin is a subunit of the plasma membrane adapter (By similarity).
Adaptins are components of the adapter complexes which link clathrin to receptors in coated vesicles. Clathrin-associated protein complexes are believed to interact with the cytoplasmic tails of membrane proteins, leading to their selection and concentration. AP-2alpha is a subunit of the plasma membrane adapter.
Adaptins are components of the adaptor complexes which link clathrin to receptors in coated vesicles. Clathrin-associated protein complexes are believed to interact with the cytoplasmic tails of membrane proteins, leading to their selection and concentration.
Subunit
Adaptor protein complex 2 (AP-2) is a heterotetramer composed of two large adaptins (alpha-type and beta-type subunits), a medium adaptin (mu-type subunit) and a small adaptin (sigma-type subunit).
Adaptor protein complex 2 (AP-2) is a heterotetramer composed of two large adaptins (alpha-type and beta-type subunits), a medium adaptin (mu-type subunit AP50) and a small adaptin (sigma-type subunit AP17).
Adaptor protein complex 2 (AP-2) is a heterotetramer composed of two large adaptins (alpha-type and beta-type subunits), a medium adaptin (mu-type subunit AP50) and a small adaptin (sigma-type subunit AP17).
Similarity
Belongs to the adaptor complexes large subunit family.
Belongs to the adapter complexes large subunit family.
Belongs to the adapter complexes large subunit family.
Keywords
Cell membrane
Coated pit
Complete proteome
Endocytosis
Membrane
Protein transport
Reference proteome
Transport
Alternative splicing
Phosphoprotein
RNA editing
Feature
chain AP-2 complex subunit alpha
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9J7H4
A0A2W1BXL5
A0A1E1WGS0
A0A2H1V2Z5
A0A194QTZ9
A0A194PM39
+ More
A0A212EQQ2 A0A0L7LFU1 A0A1Y1KZK6 V5IA26 A0A034VTV9 N6TXW3 A0A1W4WTG7 W8BAQ1 A0A0K8VJ03 D6W7L4 A0A0A1XAN1 A0A0C9RDP5 A0A336KC15 A0A336M3M0 A0A1B6F6F6 A0A1I8QF67 A0A1A9YSF6 A0A1I8MJ14 A0A0L0CCN8 U5EX15 A0A182GJ46 A0A1B0FD53 A0A1A9V0X6 A0A1A9ZQQ7 Q17CP2 A0A151IK21 A0A3L8E6I8 A0A151I5J6 A0A158NWA4 F4WLP8 A0A195EC81 A0A2M4BCJ2 E2A8U3 A0A151JTU0 A0A232FFM4 B4G8A5 Q29N38 A0A182F4E2 W5J6P4 A0A182NE80 A0A2M4ACF8 A0A1A9WUR4 A0A1W4UQ49 A0A3B0K163 A0A0J7KSA5 A0A0M4E743 A0A182Y5R8 B4N0V9 A0A182RGH4 A0A182VV19 A0A182Q353 B4LTK9 A0A1Q3FAY3 A0A1Q3FAR8 B4ICW5 A0A1L8DVR8 A0A182MDW3 Q7QG73 A0A1B6DVE5 B3N7P9 B4Q635 P91926 A0A182JGX0 B3MUI1 B4P2D9 B0XEL0 A0A182JYE8 A0A1J1J1F9 B4KFJ2 A0A182P954 A0A088APJ2 B4JDP0 A0A0A9YVV3 A0A069DXV4 A0A0K8V223 A0A2A3EMD2 A0A0V0G7Q7 A0A023F5Q8 A0A182UBX7 A0A151X1G5 A0A0M9A3D5 A0A182VNP6 P91926-2 E8NH12 A0A084VPB0 A0A310SAK0 A0A182IE13 A0A2H8TLI9 A0A2S2PGU7 X1XNU7 A0A182T5A2 A0A0P4WDI2 T1IZY9 A0A0P5MQY6
A0A212EQQ2 A0A0L7LFU1 A0A1Y1KZK6 V5IA26 A0A034VTV9 N6TXW3 A0A1W4WTG7 W8BAQ1 A0A0K8VJ03 D6W7L4 A0A0A1XAN1 A0A0C9RDP5 A0A336KC15 A0A336M3M0 A0A1B6F6F6 A0A1I8QF67 A0A1A9YSF6 A0A1I8MJ14 A0A0L0CCN8 U5EX15 A0A182GJ46 A0A1B0FD53 A0A1A9V0X6 A0A1A9ZQQ7 Q17CP2 A0A151IK21 A0A3L8E6I8 A0A151I5J6 A0A158NWA4 F4WLP8 A0A195EC81 A0A2M4BCJ2 E2A8U3 A0A151JTU0 A0A232FFM4 B4G8A5 Q29N38 A0A182F4E2 W5J6P4 A0A182NE80 A0A2M4ACF8 A0A1A9WUR4 A0A1W4UQ49 A0A3B0K163 A0A0J7KSA5 A0A0M4E743 A0A182Y5R8 B4N0V9 A0A182RGH4 A0A182VV19 A0A182Q353 B4LTK9 A0A1Q3FAY3 A0A1Q3FAR8 B4ICW5 A0A1L8DVR8 A0A182MDW3 Q7QG73 A0A1B6DVE5 B3N7P9 B4Q635 P91926 A0A182JGX0 B3MUI1 B4P2D9 B0XEL0 A0A182JYE8 A0A1J1J1F9 B4KFJ2 A0A182P954 A0A088APJ2 B4JDP0 A0A0A9YVV3 A0A069DXV4 A0A0K8V223 A0A2A3EMD2 A0A0V0G7Q7 A0A023F5Q8 A0A182UBX7 A0A151X1G5 A0A0M9A3D5 A0A182VNP6 P91926-2 E8NH12 A0A084VPB0 A0A310SAK0 A0A182IE13 A0A2H8TLI9 A0A2S2PGU7 X1XNU7 A0A182T5A2 A0A0P4WDI2 T1IZY9 A0A0P5MQY6
Pubmed
19121390
28756777
26354079
22118469
26227816
28004739
+ More
25348373 23537049 24495485 18362917 19820115 25830018 25315136 26108605 26483478 17510324 30249741 21347285 21719571 20798317 28648823 17994087 15632085 20920257 23761445 25244985 18057021 12364791 22936249 9118220 9285813 10731132 12537572 17018572 18327897 17550304 25401762 26334808 25474469 24438588
25348373 23537049 24495485 18362917 19820115 25830018 25315136 26108605 26483478 17510324 30249741 21347285 21719571 20798317 28648823 17994087 15632085 20920257 23761445 25244985 18057021 12364791 22936249 9118220 9285813 10731132 12537572 17018572 18327897 17550304 25401762 26334808 25474469 24438588
EMBL
BABH01013842
KZ149904
PZC78395.1
GDQN01004875
JAT86179.1
ODYU01000234
+ More
SOQ34654.1 KQ461154 KPJ08470.1 KQ459599 KPI94392.1 AGBW02013261 OWR43805.1 JTDY01001362 KOB74076.1 GEZM01075037 GEZM01075036 JAV64327.1 GALX01001920 JAB66546.1 GAKP01013405 GAKP01013404 GAKP01013403 JAC45547.1 APGK01047112 KB741077 KB632384 ENN74140.1 ERL94271.1 GAMC01012452 GAMC01012451 GAMC01012450 JAB94103.1 GDHF01016326 GDHF01013774 JAI35988.1 JAI38540.1 KQ971307 EFA11263.1 GBXI01015587 GBXI01006554 GBXI01005354 JAC98704.1 JAD07738.1 JAD08938.1 GBYB01005101 JAG74868.1 UFQS01000285 UFQT01000285 SSX02434.1 SSX22809.1 UFQS01000393 UFQT01000393 SSX03566.1 SSX23931.1 GECZ01024098 JAS45671.1 JRES01000561 KNC30198.1 GANO01001170 JAB58701.1 JXUM01054903 JXUM01054904 JXUM01067295 JXUM01067296 JXUM01067297 KQ562443 KQ561842 KXJ75904.1 KXJ77374.1 CCAG010003896 CH477305 EAT44154.1 KQ977282 KYN03973.1 QOIP01000001 RLU27589.1 KQ976434 KYM87176.1 ADTU01002959 GL888212 EGI64876.1 KQ979074 KYN22830.1 GGFJ01001624 MBW50765.1 GL437663 EFN70172.1 KQ981972 KYN31649.1 NNAY01000274 OXU29566.1 CH479180 EDW28585.1 CH379060 EAL33505.1 ADMH02002133 ETN58439.1 GGFK01005119 MBW38440.1 OUUW01000004 SPP79699.1 LBMM01003688 KMQ93233.1 CP012523 ALC38515.1 CH963920 EDW77722.1 AXCN02000868 CH940649 EDW64982.1 KRF81968.1 GFDL01010373 JAV24672.1 GFDL01010426 JAV24619.1 CH480829 EDW45391.1 GFDF01003557 JAV10527.1 AXCM01000251 AAAB01008839 GEDC01020541 GEDC01014060 GEDC01012641 GEDC01007658 JAS16757.1 JAS23238.1 JAS24657.1 JAS29640.1 CH954177 EDV57225.1 CM000361 CM002910 EDX03219.1 KMY87307.1 Y11104 Y13092 AE014134 BT032839 BT003458 CH902624 EDV33510.1 CM000157 EDW87135.1 DS232843 EDS26071.1 CVRI01000065 CRL05770.1 CH933807 EDW13107.1 KRG03580.1 CH916368 EDW03410.1 GBHO01009919 JAG33685.1 GBGD01000347 JAC88542.1 GDHF01019674 JAI32640.1 KZ288219 PBC32319.1 GECL01002791 JAP03333.1 GBBI01002203 JAC16509.1 KQ982600 KYQ54033.1 KQ435750 KOX76387.1 BT125866 ADU79254.1 ATLV01014994 KE524999 KFB39804.1 KQ762237 OAD56004.1 APCN01004553 GFXV01003212 MBW15017.1 GGMR01016044 MBY28663.1 ABLF02023931 ABLF02023933 ABLF02023934 GDRN01066780 JAI64473.1 JH431728 GDIQ01175394 JAK76331.1
SOQ34654.1 KQ461154 KPJ08470.1 KQ459599 KPI94392.1 AGBW02013261 OWR43805.1 JTDY01001362 KOB74076.1 GEZM01075037 GEZM01075036 JAV64327.1 GALX01001920 JAB66546.1 GAKP01013405 GAKP01013404 GAKP01013403 JAC45547.1 APGK01047112 KB741077 KB632384 ENN74140.1 ERL94271.1 GAMC01012452 GAMC01012451 GAMC01012450 JAB94103.1 GDHF01016326 GDHF01013774 JAI35988.1 JAI38540.1 KQ971307 EFA11263.1 GBXI01015587 GBXI01006554 GBXI01005354 JAC98704.1 JAD07738.1 JAD08938.1 GBYB01005101 JAG74868.1 UFQS01000285 UFQT01000285 SSX02434.1 SSX22809.1 UFQS01000393 UFQT01000393 SSX03566.1 SSX23931.1 GECZ01024098 JAS45671.1 JRES01000561 KNC30198.1 GANO01001170 JAB58701.1 JXUM01054903 JXUM01054904 JXUM01067295 JXUM01067296 JXUM01067297 KQ562443 KQ561842 KXJ75904.1 KXJ77374.1 CCAG010003896 CH477305 EAT44154.1 KQ977282 KYN03973.1 QOIP01000001 RLU27589.1 KQ976434 KYM87176.1 ADTU01002959 GL888212 EGI64876.1 KQ979074 KYN22830.1 GGFJ01001624 MBW50765.1 GL437663 EFN70172.1 KQ981972 KYN31649.1 NNAY01000274 OXU29566.1 CH479180 EDW28585.1 CH379060 EAL33505.1 ADMH02002133 ETN58439.1 GGFK01005119 MBW38440.1 OUUW01000004 SPP79699.1 LBMM01003688 KMQ93233.1 CP012523 ALC38515.1 CH963920 EDW77722.1 AXCN02000868 CH940649 EDW64982.1 KRF81968.1 GFDL01010373 JAV24672.1 GFDL01010426 JAV24619.1 CH480829 EDW45391.1 GFDF01003557 JAV10527.1 AXCM01000251 AAAB01008839 GEDC01020541 GEDC01014060 GEDC01012641 GEDC01007658 JAS16757.1 JAS23238.1 JAS24657.1 JAS29640.1 CH954177 EDV57225.1 CM000361 CM002910 EDX03219.1 KMY87307.1 Y11104 Y13092 AE014134 BT032839 BT003458 CH902624 EDV33510.1 CM000157 EDW87135.1 DS232843 EDS26071.1 CVRI01000065 CRL05770.1 CH933807 EDW13107.1 KRG03580.1 CH916368 EDW03410.1 GBHO01009919 JAG33685.1 GBGD01000347 JAC88542.1 GDHF01019674 JAI32640.1 KZ288219 PBC32319.1 GECL01002791 JAP03333.1 GBBI01002203 JAC16509.1 KQ982600 KYQ54033.1 KQ435750 KOX76387.1 BT125866 ADU79254.1 ATLV01014994 KE524999 KFB39804.1 KQ762237 OAD56004.1 APCN01004553 GFXV01003212 MBW15017.1 GGMR01016044 MBY28663.1 ABLF02023931 ABLF02023933 ABLF02023934 GDRN01066780 JAI64473.1 JH431728 GDIQ01175394 JAK76331.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000037510
UP000019118
+ More
UP000030742 UP000192223 UP000007266 UP000095300 UP000092443 UP000095301 UP000037069 UP000069940 UP000249989 UP000092444 UP000078200 UP000092445 UP000008820 UP000078542 UP000279307 UP000078540 UP000005205 UP000007755 UP000078492 UP000000311 UP000078541 UP000215335 UP000008744 UP000001819 UP000069272 UP000000673 UP000075884 UP000091820 UP000192221 UP000268350 UP000036403 UP000092553 UP000076408 UP000007798 UP000075900 UP000075920 UP000075886 UP000008792 UP000001292 UP000075883 UP000007062 UP000008711 UP000000304 UP000000803 UP000075880 UP000007801 UP000002282 UP000002320 UP000075881 UP000183832 UP000009192 UP000075885 UP000005203 UP000001070 UP000242457 UP000075902 UP000075809 UP000053105 UP000075903 UP000030765 UP000075840 UP000007819 UP000075901
UP000030742 UP000192223 UP000007266 UP000095300 UP000092443 UP000095301 UP000037069 UP000069940 UP000249989 UP000092444 UP000078200 UP000092445 UP000008820 UP000078542 UP000279307 UP000078540 UP000005205 UP000007755 UP000078492 UP000000311 UP000078541 UP000215335 UP000008744 UP000001819 UP000069272 UP000000673 UP000075884 UP000091820 UP000192221 UP000268350 UP000036403 UP000092553 UP000076408 UP000007798 UP000075900 UP000075920 UP000075886 UP000008792 UP000001292 UP000075883 UP000007062 UP000008711 UP000000304 UP000000803 UP000075880 UP000007801 UP000002282 UP000002320 UP000075881 UP000183832 UP000009192 UP000075885 UP000005203 UP000001070 UP000242457 UP000075902 UP000075809 UP000053105 UP000075903 UP000030765 UP000075840 UP000007819 UP000075901
Interpro
IPR009028
Coatomer/calthrin_app_sub_C
+ More
IPR008152 Clathrin_a/b/g-adaptin_app_Ig
IPR011989 ARM-like
IPR017104 AP2_complex_asu
IPR013041 Clathrin_app_Ig-like_sf
IPR002553 Clathrin/coatomer_adapt-like_N
IPR003164 Clathrin_a-adaptin_app_sub_C
IPR016024 ARM-type_fold
IPR012295 TBP_dom_sf
IPR002327 Cyt_c_1A/1B
IPR036909 Cyt_c-like_dom_sf
IPR009056 Cyt_c-like_dom
IPR008152 Clathrin_a/b/g-adaptin_app_Ig
IPR011989 ARM-like
IPR017104 AP2_complex_asu
IPR013041 Clathrin_app_Ig-like_sf
IPR002553 Clathrin/coatomer_adapt-like_N
IPR003164 Clathrin_a-adaptin_app_sub_C
IPR016024 ARM-type_fold
IPR012295 TBP_dom_sf
IPR002327 Cyt_c_1A/1B
IPR036909 Cyt_c-like_dom_sf
IPR009056 Cyt_c-like_dom
Gene 3D
ProteinModelPortal
H9J7H4
A0A2W1BXL5
A0A1E1WGS0
A0A2H1V2Z5
A0A194QTZ9
A0A194PM39
+ More
A0A212EQQ2 A0A0L7LFU1 A0A1Y1KZK6 V5IA26 A0A034VTV9 N6TXW3 A0A1W4WTG7 W8BAQ1 A0A0K8VJ03 D6W7L4 A0A0A1XAN1 A0A0C9RDP5 A0A336KC15 A0A336M3M0 A0A1B6F6F6 A0A1I8QF67 A0A1A9YSF6 A0A1I8MJ14 A0A0L0CCN8 U5EX15 A0A182GJ46 A0A1B0FD53 A0A1A9V0X6 A0A1A9ZQQ7 Q17CP2 A0A151IK21 A0A3L8E6I8 A0A151I5J6 A0A158NWA4 F4WLP8 A0A195EC81 A0A2M4BCJ2 E2A8U3 A0A151JTU0 A0A232FFM4 B4G8A5 Q29N38 A0A182F4E2 W5J6P4 A0A182NE80 A0A2M4ACF8 A0A1A9WUR4 A0A1W4UQ49 A0A3B0K163 A0A0J7KSA5 A0A0M4E743 A0A182Y5R8 B4N0V9 A0A182RGH4 A0A182VV19 A0A182Q353 B4LTK9 A0A1Q3FAY3 A0A1Q3FAR8 B4ICW5 A0A1L8DVR8 A0A182MDW3 Q7QG73 A0A1B6DVE5 B3N7P9 B4Q635 P91926 A0A182JGX0 B3MUI1 B4P2D9 B0XEL0 A0A182JYE8 A0A1J1J1F9 B4KFJ2 A0A182P954 A0A088APJ2 B4JDP0 A0A0A9YVV3 A0A069DXV4 A0A0K8V223 A0A2A3EMD2 A0A0V0G7Q7 A0A023F5Q8 A0A182UBX7 A0A151X1G5 A0A0M9A3D5 A0A182VNP6 P91926-2 E8NH12 A0A084VPB0 A0A310SAK0 A0A182IE13 A0A2H8TLI9 A0A2S2PGU7 X1XNU7 A0A182T5A2 A0A0P4WDI2 T1IZY9 A0A0P5MQY6
A0A212EQQ2 A0A0L7LFU1 A0A1Y1KZK6 V5IA26 A0A034VTV9 N6TXW3 A0A1W4WTG7 W8BAQ1 A0A0K8VJ03 D6W7L4 A0A0A1XAN1 A0A0C9RDP5 A0A336KC15 A0A336M3M0 A0A1B6F6F6 A0A1I8QF67 A0A1A9YSF6 A0A1I8MJ14 A0A0L0CCN8 U5EX15 A0A182GJ46 A0A1B0FD53 A0A1A9V0X6 A0A1A9ZQQ7 Q17CP2 A0A151IK21 A0A3L8E6I8 A0A151I5J6 A0A158NWA4 F4WLP8 A0A195EC81 A0A2M4BCJ2 E2A8U3 A0A151JTU0 A0A232FFM4 B4G8A5 Q29N38 A0A182F4E2 W5J6P4 A0A182NE80 A0A2M4ACF8 A0A1A9WUR4 A0A1W4UQ49 A0A3B0K163 A0A0J7KSA5 A0A0M4E743 A0A182Y5R8 B4N0V9 A0A182RGH4 A0A182VV19 A0A182Q353 B4LTK9 A0A1Q3FAY3 A0A1Q3FAR8 B4ICW5 A0A1L8DVR8 A0A182MDW3 Q7QG73 A0A1B6DVE5 B3N7P9 B4Q635 P91926 A0A182JGX0 B3MUI1 B4P2D9 B0XEL0 A0A182JYE8 A0A1J1J1F9 B4KFJ2 A0A182P954 A0A088APJ2 B4JDP0 A0A0A9YVV3 A0A069DXV4 A0A0K8V223 A0A2A3EMD2 A0A0V0G7Q7 A0A023F5Q8 A0A182UBX7 A0A151X1G5 A0A0M9A3D5 A0A182VNP6 P91926-2 E8NH12 A0A084VPB0 A0A310SAK0 A0A182IE13 A0A2H8TLI9 A0A2S2PGU7 X1XNU7 A0A182T5A2 A0A0P4WDI2 T1IZY9 A0A0P5MQY6
PDB
2XA7
E-value=0,
Score=2707
Ontologies
GO
GO:0072583
GO:0006886
GO:0035615
GO:0030122
GO:0006897
GO:0005905
GO:0005737
GO:0048488
GO:0005886
GO:0015031
GO:0098793
GO:0008356
GO:0030100
GO:0045807
GO:0008104
GO:0010508
GO:1990386
GO:0008565
GO:0030707
GO:0048489
GO:0009055
GO:0046872
GO:0020037
GO:0016192
GO:0030131
GO:0030117
GO:0005488
GO:0016020
GO:0016742
GO:0003824
Topology
Subcellular location
Cytoplasm
Membrane
Coated pit
Cell membrane Component of the coat surrounding the cytoplasmic face of coated vesicles in the plasma membrane. With evidence from 2 publications.
Membrane
Coated pit
Cell membrane Component of the coat surrounding the cytoplasmic face of coated vesicles in the plasma membrane. With evidence from 2 publications.
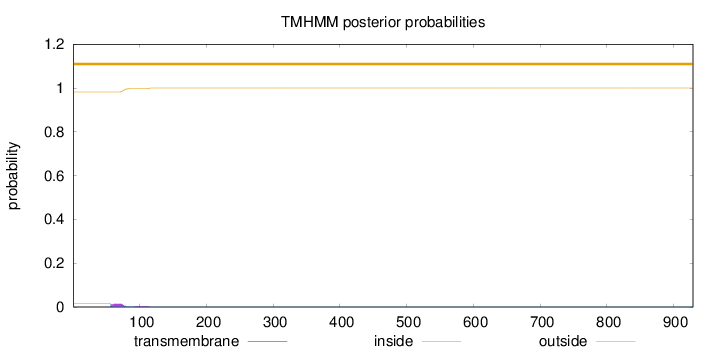
Length:
929
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.35419
Exp number, first 60 AAs:
0.04975
Total prob of N-in:
0.01762
outside
1 - 929
Population Genetic Test Statistics
Pi
5.349285
Theta
18.649393
Tajima's D
-2.04379
CLR
1462.922816
CSRT
0.0121993900304985
Interpretation
Uncertain
