Gene
KWMTBOMO04540
Pre Gene Modal
BGIBMGA005464
Annotation
PREDICTED:_trafficking_protein_particle_complex_subunit_13-like_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.834 Nuclear Reliability : 1.562
Sequence
CDS
ATGGATCCAAAAGAAGCGGTTGAACATTTAGTTGCTCTGAAGGTGATGAGACTTACAAAGCCAGCATTGATAAGCCCTAATATCGTCACTTGTGACTCAAAAGATTTGCCAGGTAATATTCTCAATAATTATCTAAAAGATGATGCCACAGCAGTCAGTCAAATGGAAACTCTTGCTGCTGGACAATTCTTACTACTTCCTCAAAGTTTTGGAAATATTTATTTAGGGGAAACATTCTCTTGTTATGTTTGTGTTCACAATGAAACGAATCAACCTGTACAAAGTGTCTCCATAATAACTAGTCTACAAACTAATTCACAAAGAATAGAGCTTACAGCACAGGAGGGAACTACCCCTACTATGTTGGATGTTGATGAAACCCTTAGTGATGTTATTCACCATGAGGTCAAAGATTTAGGAACACATATATTAGTATGTGAAGTTTCGTACATGTCCAACTACAATACGTTAGCATCGTTCCGGAAATTCTTCAAATTTGAAGTCATGAAACCACTAGATGTGAAAACTAAATTTTATAATGCTGAATCAGGGGATATTTATCTTGAAGCACAAGTTCACAACATAACATCTGGGCCCATTATATTTGAGCAAGTATCTTTGGAAAGTTCTCAGCAATTCACTGTAAGTTCTTTGAATGAAGTGAATGGTGAGTCAGTGTTTGGTGAAGTGACATTATTACAGCCACAAGAGAGCTGTCAGTTCTTGTACTGGCTTAAACCAAAGGAAAACATGGCAAAAGAAGTCAAACCACATGGCCCTATGAGAAATATTGGAAAACTAGATATAGTCTGGAGGTCTAACTTAGGTGAGAAAGGCCGATTACAGACTAGTCAGTTGGAAGCCATGGCACTCGATCATGACGACTTAAGACTGACAGTTGAACAAATTCCTAGTAAAGTATGTGCAGATGAAGCTTTCACATTTAAATGTAAAGTTGTGAATGCCAGTGAAAGAACTTTAGATCTCATATTAAAACTGCGTTCTCCGCAAGACTCTAGTCTCCTATGGTGCGGAATTTCTAACAGAAAATTAGGGCCTCTAGAACCCGGAAAGACAACATTCATAAATTTAACTGCTTTACCAATCAACACAGGCTTACAGAAAGTGACTGGTGTGTCACTTGTTGATTTGTTTTTAAAACGCACTTATGATTATGATCATTTAGCTTCAGTTTATGTATATTAA
Protein
MDPKEAVEHLVALKVMRLTKPALISPNIVTCDSKDLPGNILNNYLKDDATAVSQMETLAAGQFLLLPQSFGNIYLGETFSCYVCVHNETNQPVQSVSIITSLQTNSQRIELTAQEGTTPTMLDVDETLSDVIHHEVKDLGTHILVCEVSYMSNYNTLASFRKFFKFEVMKPLDVKTKFYNAESGDIYLEAQVHNITSGPIIFEQVSLESSQQFTVSSLNEVNGESVFGEVTLLQPQESCQFLYWLKPKENMAKEVKPHGPMRNIGKLDIVWRSNLGEKGRLQTSQLEAMALDHDDLRLTVEQIPSKVCADEAFTFKCKVVNASERTLDLILKLRSPQDSSLLWCGISNRKLGPLEPGKTTFINLTALPINTGLQKVTGVSLVDLFLKRTYDYDHLASVYVY
Summary
Uniprot
H9J7H2
A0A2W1BVM3
A0A2H1V1D7
A0A1E1W8F4
S4NP07
A0A0L7LUC5
+ More
A0A212EQP3 A0A194PTD3 A0A194QSK4 A0A2J7PU27 A0A067R570 A0A2J7PU29 A0A182NLQ1 A0A182QZF6 A0A1B6I1G1 D6WTU7 J3JV74 A0A084WKL9 A0A2M3ZAM0 A0A2M4BNY8 A0A2M4ARA4 A0A182FCI4 A0A0C9RNB4 A0A182IUG0 A0A182G1K2 W5JTD3 A0A182HLX3 A0A182X527 A0A182LJD5 A0A182TQ78 A0A182VE04 A0A0T6ATP0 Q7Q714 E2ATU6 A0A182Y130 A0A182PHF1 A0A182JZW0 A0A182RMH2 A0A2A3ETZ9 K7IVH3 A0A232EZF1 A0A182LWK3 A0A182WKE9 Q17NV6 B0WC50 A0A088AGY5 A0A195BN46 A0A158P014 A0A026WNU6 A0A1Y1NKT0 A0A151XJ77 F4X3N3 A0A1Q3F7L4 A0A195CQL1 A0A1W4X930 A0A154PDM6 A0A0L7RHK5 A0A1I8QEU4 E2BY98 C3ZJ54 A0A1Y1NJN0 A0A195F2Z1 A0A195E0Q6 A0A0Q3UQJ7 A0A1Q3F7K9 A0A2I0U1C2 A0A2R5L579 A0A218V918 A0A151MXF2 U3K584 A0A293LLM1 F6UA19 A0A341C2X7 A0A1I8MLW7 A0A3Q1DHM1 A0A2Y9Q3G4 B7QCD0 A0A383ZLQ2 A0A2Y9FVZ0 L5KIL3 A0A151MXJ5 I3MYS4 G3WFL4 A0A2U4AX20 A0A131Y075 A0A384C409 A0A3Q7WKG6 A0A3P8RTU8 A0A1A7WKM4 A0A1L1RS53 H0YXC0 K7FU72 A0A3Q1GTK1 A0A2J8VU20 A0A1W4Z8Y0 F1NA83 A0A3Q3GIS0 A0A3Q2C9B8 A0A1B0CU20 A0A3B3XL68
A0A212EQP3 A0A194PTD3 A0A194QSK4 A0A2J7PU27 A0A067R570 A0A2J7PU29 A0A182NLQ1 A0A182QZF6 A0A1B6I1G1 D6WTU7 J3JV74 A0A084WKL9 A0A2M3ZAM0 A0A2M4BNY8 A0A2M4ARA4 A0A182FCI4 A0A0C9RNB4 A0A182IUG0 A0A182G1K2 W5JTD3 A0A182HLX3 A0A182X527 A0A182LJD5 A0A182TQ78 A0A182VE04 A0A0T6ATP0 Q7Q714 E2ATU6 A0A182Y130 A0A182PHF1 A0A182JZW0 A0A182RMH2 A0A2A3ETZ9 K7IVH3 A0A232EZF1 A0A182LWK3 A0A182WKE9 Q17NV6 B0WC50 A0A088AGY5 A0A195BN46 A0A158P014 A0A026WNU6 A0A1Y1NKT0 A0A151XJ77 F4X3N3 A0A1Q3F7L4 A0A195CQL1 A0A1W4X930 A0A154PDM6 A0A0L7RHK5 A0A1I8QEU4 E2BY98 C3ZJ54 A0A1Y1NJN0 A0A195F2Z1 A0A195E0Q6 A0A0Q3UQJ7 A0A1Q3F7K9 A0A2I0U1C2 A0A2R5L579 A0A218V918 A0A151MXF2 U3K584 A0A293LLM1 F6UA19 A0A341C2X7 A0A1I8MLW7 A0A3Q1DHM1 A0A2Y9Q3G4 B7QCD0 A0A383ZLQ2 A0A2Y9FVZ0 L5KIL3 A0A151MXJ5 I3MYS4 G3WFL4 A0A2U4AX20 A0A131Y075 A0A384C409 A0A3Q7WKG6 A0A3P8RTU8 A0A1A7WKM4 A0A1L1RS53 H0YXC0 K7FU72 A0A3Q1GTK1 A0A2J8VU20 A0A1W4Z8Y0 F1NA83 A0A3Q3GIS0 A0A3Q2C9B8 A0A1B0CU20 A0A3B3XL68
Pubmed
19121390
28756777
23622113
26227816
22118469
26354079
+ More
24845553 18362917 19820115 22516182 24438588 26483478 20920257 23761445 20966253 12364791 14747013 17210077 20798317 25244985 20075255 28648823 17510324 21347285 24508170 30249741 28004739 21719571 18563158 22293439 17495919 25315136 23258410 21709235 15592404 20360741 17381049
24845553 18362917 19820115 22516182 24438588 26483478 20920257 23761445 20966253 12364791 14747013 17210077 20798317 25244985 20075255 28648823 17510324 21347285 24508170 30249741 28004739 21719571 18563158 22293439 17495919 25315136 23258410 21709235 15592404 20360741 17381049
EMBL
BABH01013840
KZ149904
PZC78391.1
ODYU01000234
SOQ34657.1
GDQN01007913
+ More
GDQN01000738 JAT83141.1 JAT90316.1 GAIX01012049 JAA80511.1 JTDY01000070 KOB79047.1 AGBW02013261 OWR43808.1 KQ459599 KPI94395.1 KQ461154 KPJ08467.1 NEVH01021208 PNF19837.1 KK852880 KDR14425.1 PNF19839.1 AXCN02000986 GECU01026950 JAS80756.1 KQ971352 EFA07324.1 BT127141 AEE62103.1 ATLV01024126 KE525349 KFB50763.1 GGFM01004833 MBW25584.1 GGFJ01005646 MBW54787.1 GGFK01010005 MBW43326.1 GBYB01008571 JAG78338.1 JXUM01137137 KQ568531 KXJ68972.1 ADMH02000542 ETN65994.1 APCN01004270 LJIG01022907 KRT78119.1 AAAB01008960 EAA11831.3 GL442691 EFN63179.1 KZ288189 PBC34686.1 NNAY01001524 OXU23699.1 AXCM01016355 CH477294 CH477196 EAT44427.1 EAT48368.1 DS231885 EDS43202.1 KQ976440 KYM86604.1 ADTU01005245 KK107139 QOIP01000003 EZA57732.1 RLU24983.1 GEZM01004708 JAV96327.1 KQ982074 KYQ60454.1 GL888624 EGI58830.1 GFDL01011503 JAV23542.1 KQ977394 KYN03033.1 KQ434879 KZC09941.1 KQ414591 KOC70283.1 GL451420 EFN79377.1 GG666631 EEN47385.1 GEZM01004709 JAV96326.1 KQ981856 KYN34547.1 KQ979955 KYN18517.1 LMAW01002889 KQK76010.1 GFDL01011552 JAV23493.1 KZ506399 PKU39825.1 GGLE01000507 MBY04633.1 MUZQ01000025 OWK62524.1 AKHW03004724 KYO29184.1 AGTO01020873 GFWV01002865 MAA27595.1 ABJB011052101 DS906639 EEC16502.1 KB030673 ELK11395.1 KYO29185.1 AGTP01014028 AGTP01014029 AEFK01050382 AEFK01050383 AEFK01050384 GEFM01003888 JAP71908.1 HADW01004923 HADX01007237 SBP06323.1 AC145935 AC187110 ABQF01033291 ABQF01033292 ABQF01033293 AGCU01045555 AGCU01045556 AGCU01045557 NDHI03003409 PNJ61015.1 AJWK01028329
GDQN01000738 JAT83141.1 JAT90316.1 GAIX01012049 JAA80511.1 JTDY01000070 KOB79047.1 AGBW02013261 OWR43808.1 KQ459599 KPI94395.1 KQ461154 KPJ08467.1 NEVH01021208 PNF19837.1 KK852880 KDR14425.1 PNF19839.1 AXCN02000986 GECU01026950 JAS80756.1 KQ971352 EFA07324.1 BT127141 AEE62103.1 ATLV01024126 KE525349 KFB50763.1 GGFM01004833 MBW25584.1 GGFJ01005646 MBW54787.1 GGFK01010005 MBW43326.1 GBYB01008571 JAG78338.1 JXUM01137137 KQ568531 KXJ68972.1 ADMH02000542 ETN65994.1 APCN01004270 LJIG01022907 KRT78119.1 AAAB01008960 EAA11831.3 GL442691 EFN63179.1 KZ288189 PBC34686.1 NNAY01001524 OXU23699.1 AXCM01016355 CH477294 CH477196 EAT44427.1 EAT48368.1 DS231885 EDS43202.1 KQ976440 KYM86604.1 ADTU01005245 KK107139 QOIP01000003 EZA57732.1 RLU24983.1 GEZM01004708 JAV96327.1 KQ982074 KYQ60454.1 GL888624 EGI58830.1 GFDL01011503 JAV23542.1 KQ977394 KYN03033.1 KQ434879 KZC09941.1 KQ414591 KOC70283.1 GL451420 EFN79377.1 GG666631 EEN47385.1 GEZM01004709 JAV96326.1 KQ981856 KYN34547.1 KQ979955 KYN18517.1 LMAW01002889 KQK76010.1 GFDL01011552 JAV23493.1 KZ506399 PKU39825.1 GGLE01000507 MBY04633.1 MUZQ01000025 OWK62524.1 AKHW03004724 KYO29184.1 AGTO01020873 GFWV01002865 MAA27595.1 ABJB011052101 DS906639 EEC16502.1 KB030673 ELK11395.1 KYO29185.1 AGTP01014028 AGTP01014029 AEFK01050382 AEFK01050383 AEFK01050384 GEFM01003888 JAP71908.1 HADW01004923 HADX01007237 SBP06323.1 AC145935 AC187110 ABQF01033291 ABQF01033292 ABQF01033293 AGCU01045555 AGCU01045556 AGCU01045557 NDHI03003409 PNJ61015.1 AJWK01028329
Proteomes
UP000005204
UP000037510
UP000007151
UP000053268
UP000053240
UP000235965
+ More
UP000027135 UP000075884 UP000075886 UP000007266 UP000030765 UP000069272 UP000075880 UP000069940 UP000249989 UP000000673 UP000075840 UP000076407 UP000075882 UP000075902 UP000075903 UP000007062 UP000000311 UP000076408 UP000075885 UP000075881 UP000075900 UP000242457 UP000002358 UP000215335 UP000075883 UP000075920 UP000008820 UP000002320 UP000005203 UP000078540 UP000005205 UP000053097 UP000279307 UP000075809 UP000007755 UP000078542 UP000192223 UP000076502 UP000053825 UP000095300 UP000008237 UP000001554 UP000078541 UP000078492 UP000051836 UP000197619 UP000050525 UP000016665 UP000002280 UP000252040 UP000095301 UP000257160 UP000248483 UP000001555 UP000261681 UP000248480 UP000010552 UP000005215 UP000007648 UP000245320 UP000261680 UP000286642 UP000265080 UP000000539 UP000007754 UP000007267 UP000257200 UP000192224 UP000264800 UP000265020 UP000092461 UP000261480
UP000027135 UP000075884 UP000075886 UP000007266 UP000030765 UP000069272 UP000075880 UP000069940 UP000249989 UP000000673 UP000075840 UP000076407 UP000075882 UP000075902 UP000075903 UP000007062 UP000000311 UP000076408 UP000075885 UP000075881 UP000075900 UP000242457 UP000002358 UP000215335 UP000075883 UP000075920 UP000008820 UP000002320 UP000005203 UP000078540 UP000005205 UP000053097 UP000279307 UP000075809 UP000007755 UP000078542 UP000192223 UP000076502 UP000053825 UP000095300 UP000008237 UP000001554 UP000078541 UP000078492 UP000051836 UP000197619 UP000050525 UP000016665 UP000002280 UP000252040 UP000095301 UP000257160 UP000248483 UP000001555 UP000261681 UP000248480 UP000010552 UP000005215 UP000007648 UP000245320 UP000261680 UP000286642 UP000265080 UP000000539 UP000007754 UP000007267 UP000257200 UP000192224 UP000264800 UP000265020 UP000092461 UP000261480
PRIDE
Pfam
PF06159 DUF974
Interpro
IPR010378
TRAPPC13
ProteinModelPortal
H9J7H2
A0A2W1BVM3
A0A2H1V1D7
A0A1E1W8F4
S4NP07
A0A0L7LUC5
+ More
A0A212EQP3 A0A194PTD3 A0A194QSK4 A0A2J7PU27 A0A067R570 A0A2J7PU29 A0A182NLQ1 A0A182QZF6 A0A1B6I1G1 D6WTU7 J3JV74 A0A084WKL9 A0A2M3ZAM0 A0A2M4BNY8 A0A2M4ARA4 A0A182FCI4 A0A0C9RNB4 A0A182IUG0 A0A182G1K2 W5JTD3 A0A182HLX3 A0A182X527 A0A182LJD5 A0A182TQ78 A0A182VE04 A0A0T6ATP0 Q7Q714 E2ATU6 A0A182Y130 A0A182PHF1 A0A182JZW0 A0A182RMH2 A0A2A3ETZ9 K7IVH3 A0A232EZF1 A0A182LWK3 A0A182WKE9 Q17NV6 B0WC50 A0A088AGY5 A0A195BN46 A0A158P014 A0A026WNU6 A0A1Y1NKT0 A0A151XJ77 F4X3N3 A0A1Q3F7L4 A0A195CQL1 A0A1W4X930 A0A154PDM6 A0A0L7RHK5 A0A1I8QEU4 E2BY98 C3ZJ54 A0A1Y1NJN0 A0A195F2Z1 A0A195E0Q6 A0A0Q3UQJ7 A0A1Q3F7K9 A0A2I0U1C2 A0A2R5L579 A0A218V918 A0A151MXF2 U3K584 A0A293LLM1 F6UA19 A0A341C2X7 A0A1I8MLW7 A0A3Q1DHM1 A0A2Y9Q3G4 B7QCD0 A0A383ZLQ2 A0A2Y9FVZ0 L5KIL3 A0A151MXJ5 I3MYS4 G3WFL4 A0A2U4AX20 A0A131Y075 A0A384C409 A0A3Q7WKG6 A0A3P8RTU8 A0A1A7WKM4 A0A1L1RS53 H0YXC0 K7FU72 A0A3Q1GTK1 A0A2J8VU20 A0A1W4Z8Y0 F1NA83 A0A3Q3GIS0 A0A3Q2C9B8 A0A1B0CU20 A0A3B3XL68
A0A212EQP3 A0A194PTD3 A0A194QSK4 A0A2J7PU27 A0A067R570 A0A2J7PU29 A0A182NLQ1 A0A182QZF6 A0A1B6I1G1 D6WTU7 J3JV74 A0A084WKL9 A0A2M3ZAM0 A0A2M4BNY8 A0A2M4ARA4 A0A182FCI4 A0A0C9RNB4 A0A182IUG0 A0A182G1K2 W5JTD3 A0A182HLX3 A0A182X527 A0A182LJD5 A0A182TQ78 A0A182VE04 A0A0T6ATP0 Q7Q714 E2ATU6 A0A182Y130 A0A182PHF1 A0A182JZW0 A0A182RMH2 A0A2A3ETZ9 K7IVH3 A0A232EZF1 A0A182LWK3 A0A182WKE9 Q17NV6 B0WC50 A0A088AGY5 A0A195BN46 A0A158P014 A0A026WNU6 A0A1Y1NKT0 A0A151XJ77 F4X3N3 A0A1Q3F7L4 A0A195CQL1 A0A1W4X930 A0A154PDM6 A0A0L7RHK5 A0A1I8QEU4 E2BY98 C3ZJ54 A0A1Y1NJN0 A0A195F2Z1 A0A195E0Q6 A0A0Q3UQJ7 A0A1Q3F7K9 A0A2I0U1C2 A0A2R5L579 A0A218V918 A0A151MXF2 U3K584 A0A293LLM1 F6UA19 A0A341C2X7 A0A1I8MLW7 A0A3Q1DHM1 A0A2Y9Q3G4 B7QCD0 A0A383ZLQ2 A0A2Y9FVZ0 L5KIL3 A0A151MXJ5 I3MYS4 G3WFL4 A0A2U4AX20 A0A131Y075 A0A384C409 A0A3Q7WKG6 A0A3P8RTU8 A0A1A7WKM4 A0A1L1RS53 H0YXC0 K7FU72 A0A3Q1GTK1 A0A2J8VU20 A0A1W4Z8Y0 F1NA83 A0A3Q3GIS0 A0A3Q2C9B8 A0A1B0CU20 A0A3B3XL68
Ontologies
GO
PANTHER
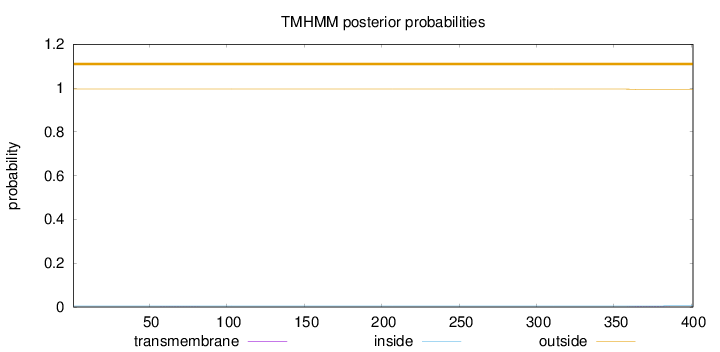
Topology
Length:
401
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04285
Exp number, first 60 AAs:
0.00152
Total prob of N-in:
0.00452
outside
1 - 401
Population Genetic Test Statistics
Pi
59.491597
Theta
126.942516
Tajima's D
-1.867312
CLR
158.640572
CSRT
0.0247487625618719
Interpretation
Uncertain
