Pre Gene Modal
BGIBMGA005344
Annotation
PREDICTED:_cytoplasmic_phosphatidylinositol_transfer_protein_1_[Papilio_machaon]
Full name
Cytoplasmic phosphatidylinositol transfer protein 1
Alternative Name
Retinal degeneration B homolog beta
Mammalian rdgB homolog beta
Mammalian rdgB homolog beta
Location in the cell
Cytoplasmic Reliability : 2.029
Sequence
CDS
ATGGTATTAACAAGAGAATACAGAATATGCATGCCTATGACTGTAGAAGAGTACCGTATTGGTCAATTATACATGATAGCAAGACACAGTTTTGAGCAATCTAGTAATGGTGAAGGTGTGGAGGTGATAGCAAATGAACAGGTGAATGATGAAAAAAATGGTATAGGACAGTTCACTGAGAAGAGAATCCATTTATCTAGTCATTTGCCTTATTGGGTCCAATCTATGATACCAAAAATATTTTATGTTACGGAAAAGGCATGGAATTTTTACCCATACACAATAACCGAATACACATGCTCATTCATACCGAAATTTTCCATCTCTATTCAAACAAGGTATGAAGATAATAATGGGACCACTGAAAATTGCCTTGGTTTCACCTCTGAAGAGTTAGAACAGCGAGAAGTAGACTTTATAGACATTGCATATGATGATATCAAATCACACCATTATAAAGAAGCTGAAGACCCTAAGTTTTTCAAATCAGAGAAAACCGGCCGCGGACCTCTAATTGAAGGGTGGAGGGACACATTCAAACCCATAATGTGTTGTTACAAAGCTGTTTCTATTAAGTTTGAAGTATGGGGCTTGCAAACCAGAGTTGAAGAGTATGTGCAGGGAGCAATTCGCGAAATTTTATTATTGGGACATCGTCAGGCATTTGCCTGGATGGATGACTGGTATAACATGAGTATAGAAGACGTAAGAGAATATGAACGCGAAATGCAAGCCAAGACGAATATAAAGTTGAAAACGACTTCTGAGAATGGTGAAGGGAAATCTGACGAGAGTGACGAGACGAAATCGAATTCTTCGAAACCACAGACTCCGAGAACTCCGAAATCGCCAAAGAGTCCAAGCAGTACTCCGACAGCAGAGGGACGAAGTTGGTTCAATTGGTCGTAA
Protein
MVLTREYRICMPMTVEEYRIGQLYMIARHSFEQSSNGEGVEVIANEQVNDEKNGIGQFTEKRIHLSSHLPYWVQSMIPKIFYVTEKAWNFYPYTITEYTCSFIPKFSISIQTRYEDNNGTTENCLGFTSEELEQREVDFIDIAYDDIKSHHYKEAEDPKFFKSEKTGRGPLIEGWRDTFKPIMCCYKAVSIKFEVWGLQTRVEEYVQGAIREILLLGHRQAFAWMDDWYNMSIEDVREYEREMQAKTNIKLKTTSENGEGKSDESDETKSNSSKPQTPRTPKSPKSPSSTPTAEGRSWFNWS
Summary
Description
Phosphatidylinositol transfer proteins mediate the monomeric transport of lipids by shielding a lipid from the aqueous environment and binding the lipid in a hydrophobic cavity.
Phosphatidylinositol transfer proteins mediate the monomeric transport of lipids by shielding a lipid from the aqueous environment and binding the lipid in a hydrophobic cavity. Able to transfer phosphatidylinositol in vitro. Isoform 2 specifically binds to phosphatidylinositol but not to other phospholipids and may play a role in the phosphoinositide-mediated signaling in the neural development.
Phosphatidylinositol transfer proteins mediate the monomeric transport of lipids by shielding a lipid from the aqueous environment and binding the lipid in a hydrophobic cavity. Able to transfer phosphatidylinositol in vitro. Isoform 2 specifically binds to phosphatidylinositol but not to other phospholipids and may play a role in the phosphoinositide-mediated signaling in the neural development.
Similarity
Belongs to the PtdIns transfer protein family. PI transfer class IIB subfamily.
Keywords
Complete proteome
Lipid transport
Lipid-binding
Reference proteome
Transport
Alternative splicing
Cytoplasm
Nucleus
Phosphoprotein
Feature
chain Cytoplasmic phosphatidylinositol transfer protein 1
splice variant In isoform 4.
splice variant In isoform 4.
Uniprot
A0A2W1BZV5
A0A2A4IYV1
A0A2H1V1D9
A0A194QY14
A0A3S2M504
A0A212EQQ6
+ More
H9J752 A0A1E1VY14 A0A0C9QHL5 E9I8B4 V9IFY7 A0A0J7NV44 A0A087ZP11 A0A026VXE8 E2BF69 A0A195BJ38 A0A195DV06 A0A195F4N3 A0A158NJU3 D6WY44 A0A3L8DNS6 A0A0L0BNY7 J3JWK5 A0A1B0D981 U4U0X8 A0A1S3D367 A0A310SMK7 A0A1Y1L4M8 A0A336LZK0 A0A151XE44 A0A067RMZ4 B3NML8 B4J8L5 A0A1B6KBC2 A0A195CL38 A0A2J7RDC4 B4MPN9 A0A1B6ISQ8 B4KMX4 J9JMS4 B4LNC4 A0A1W4XBN7 A0A0M4EDS6 A0A0F7VK74 A0A1B6GT25 A0A0M8ZXX1 A0A154NY03 A0A0J9RFI1 A0A336LXC2 D3TP06 B4HMZ1 A0A224XHE9 A0A1L8DYT7 Q28Z48 A0A0V0GBI8 A0A1B6C760 B4GIA4 A0A023F4V1 A0A2M3ZC66 A0A2M4AXX7 A0A0K8UW46 A0A069DQW1 A0A1W4VCL4 B4P554 B3MIE3 W5JDA8 Q16R16 T1PGZ5 Q9U9P7 A0A182NSG3 A0A1S4FS70 A0A182PTC3 A0A182XFC5 A0A226DBQ2 A0A3B0JQ37 A0A0A1X9P8 A0A023ELS9 A0A1I8PZW0 A0A182JTZ5 A0A0A9YYI6 A0A1Q3FDX7 L8BSU6 Q5TSY2 A0A0P6JH43 A0A1S2ZTG3 K9IVU7 A0A1S3PJB2 I3N7H0 A0A286XCN4 A0A1B0FHV0 A0A2Y9DH67 Q8K4R4-2 K9IX44 A0A3B3RWG1 G1QL61 A0A2J8V029 A0A3Q7RY60 M3VY39 A0A2Y9KR92 M3YTB3
H9J752 A0A1E1VY14 A0A0C9QHL5 E9I8B4 V9IFY7 A0A0J7NV44 A0A087ZP11 A0A026VXE8 E2BF69 A0A195BJ38 A0A195DV06 A0A195F4N3 A0A158NJU3 D6WY44 A0A3L8DNS6 A0A0L0BNY7 J3JWK5 A0A1B0D981 U4U0X8 A0A1S3D367 A0A310SMK7 A0A1Y1L4M8 A0A336LZK0 A0A151XE44 A0A067RMZ4 B3NML8 B4J8L5 A0A1B6KBC2 A0A195CL38 A0A2J7RDC4 B4MPN9 A0A1B6ISQ8 B4KMX4 J9JMS4 B4LNC4 A0A1W4XBN7 A0A0M4EDS6 A0A0F7VK74 A0A1B6GT25 A0A0M8ZXX1 A0A154NY03 A0A0J9RFI1 A0A336LXC2 D3TP06 B4HMZ1 A0A224XHE9 A0A1L8DYT7 Q28Z48 A0A0V0GBI8 A0A1B6C760 B4GIA4 A0A023F4V1 A0A2M3ZC66 A0A2M4AXX7 A0A0K8UW46 A0A069DQW1 A0A1W4VCL4 B4P554 B3MIE3 W5JDA8 Q16R16 T1PGZ5 Q9U9P7 A0A182NSG3 A0A1S4FS70 A0A182PTC3 A0A182XFC5 A0A226DBQ2 A0A3B0JQ37 A0A0A1X9P8 A0A023ELS9 A0A1I8PZW0 A0A182JTZ5 A0A0A9YYI6 A0A1Q3FDX7 L8BSU6 Q5TSY2 A0A0P6JH43 A0A1S2ZTG3 K9IVU7 A0A1S3PJB2 I3N7H0 A0A286XCN4 A0A1B0FHV0 A0A2Y9DH67 Q8K4R4-2 K9IX44 A0A3B3RWG1 G1QL61 A0A2J8V029 A0A3Q7RY60 M3VY39 A0A2Y9KR92 M3YTB3
Pubmed
28756777
26354079
22118469
19121390
21282665
24508170
+ More
20798317 21347285 18362917 19820115 30249741 26108605 22516182 23537049 28004739 24845553 17994087 22936249 20353571 15632085 25474469 26334808 17550304 20920257 23761445 17510324 25315136 10731132 12537572 10731138 25830018 24945155 25401762 12364791 14747013 17210077 21993624 12562526 16141072 19468303 15489334 15345747 21183079 29240929 17975172
20798317 21347285 18362917 19820115 30249741 26108605 22516182 23537049 28004739 24845553 17994087 22936249 20353571 15632085 25474469 26334808 17550304 20920257 23761445 17510324 25315136 10731132 12537572 10731138 25830018 24945155 25401762 12364791 14747013 17210077 21993624 12562526 16141072 19468303 15489334 15345747 21183079 29240929 17975172
EMBL
KZ149904
PZC78390.1
NWSH01005209
PCG64350.1
ODYU01000234
SOQ34658.1
+ More
KQ461154 KPJ08466.1 RSAL01000032 RVE51541.1 AGBW02013261 OWR43809.1 BABH01013837 GDQN01011475 GDQN01004953 JAT79579.1 JAT86101.1 GBYB01003019 JAG72786.1 GL761538 EFZ23142.1 JR045483 AEY59990.1 LBMM01001467 KMQ96265.1 KK107681 EZA48101.1 GL447952 EFN85655.1 KQ976465 KYM84349.1 KQ980304 KYN16745.1 KQ981820 KYN35346.1 ADTU01018383 KQ971362 EFA07896.1 QOIP01000006 RLU22100.1 JRES01001589 KNC21688.1 APGK01035766 BT127623 KB740928 AEE62585.1 ENN77950.1 AJVK01004273 AJVK01004274 KB631763 ERL85933.1 KQ762329 OAD55966.1 GEZM01064783 JAV68642.1 UFQT01000257 SSX22451.1 KQ982254 KYQ58590.1 KK852528 KDR21990.1 CH954179 EDV54957.1 CH916367 EDW01282.1 GEBQ01031237 JAT08740.1 KQ977634 KYN01172.1 NEVH01005291 PNF38837.1 CH963849 EDW74078.1 GECU01017769 JAS89937.1 CH933808 EDW09896.1 ABLF02034549 ABLF02034555 ABLF02034557 ABLF02045036 CH940648 EDW61076.1 CP012524 ALC41188.1 LN830736 CFW94245.1 GECZ01004266 JAS65503.1 KQ435798 KOX73370.1 KQ434781 KZC04497.1 CM002911 KMY94656.1 SSX22450.1 EZ423158 ADD19434.1 CH480816 EDW48341.1 GFTR01004511 JAW11915.1 GFDF01002488 JAV11596.1 CM000071 EAL25767.2 GECL01000782 JAP05342.1 GEDC01027996 GEDC01022797 JAS09302.1 JAS14501.1 CH479183 EDW36224.1 GBBI01002698 JAC16014.1 GGFM01005340 MBW26091.1 GGFK01012274 MBW45595.1 GDHF01021405 JAI30909.1 GBGD01002441 JAC86448.1 CM000158 EDW91755.1 CH902619 EDV36991.1 ADMH02001863 ETN60799.1 CH477726 EAT36858.1 KA648077 AFP62706.1 AE013599 AF160934 LNIX01000025 OXA42603.1 OUUW01000001 SPP73268.1 GBXI01006909 JAD07383.1 GAPW01003375 JAC10223.1 GBHO01032889 GBHO01032883 GBHO01005527 GBHO01005524 GBRD01017016 JAG10715.1 JAG10721.1 JAG38077.1 JAG38080.1 JAG48811.1 GFDL01009282 JAV25763.1 HF549287 CCO62048.1 AAAB01008898 EAL40545.4 GEBF01000357 JAO03276.1 GACC01000052 JAA53755.1 AGTP01052648 AGTP01052649 AGTP01052650 AGTP01052651 AGTP01052652 AGTP01052653 AGTP01052654 AGTP01052655 AGTP01052656 AGTP01052657 AAKN02045239 AAKN02045240 AAKN02045241 CCAG010015795 AB077281 AB077282 AK153625 AK171340 AL596116 AL645687 AL645905 BC108351 GABZ01007299 JAA46226.1 ADFV01091842 ADFV01091843 ADFV01091844 ADFV01091845 ADFV01091846 ADFV01091847 ADFV01091848 ADFV01091849 ADFV01091850 ADFV01091851 NDHI03003438 PNJ50874.1 AANG04004184 AEYP01025685 AEYP01025686 AEYP01025687 AEYP01025688 AEYP01025689 AEYP01025690 AEYP01025691 AEYP01025692 AEYP01025693 AEYP01025694
KQ461154 KPJ08466.1 RSAL01000032 RVE51541.1 AGBW02013261 OWR43809.1 BABH01013837 GDQN01011475 GDQN01004953 JAT79579.1 JAT86101.1 GBYB01003019 JAG72786.1 GL761538 EFZ23142.1 JR045483 AEY59990.1 LBMM01001467 KMQ96265.1 KK107681 EZA48101.1 GL447952 EFN85655.1 KQ976465 KYM84349.1 KQ980304 KYN16745.1 KQ981820 KYN35346.1 ADTU01018383 KQ971362 EFA07896.1 QOIP01000006 RLU22100.1 JRES01001589 KNC21688.1 APGK01035766 BT127623 KB740928 AEE62585.1 ENN77950.1 AJVK01004273 AJVK01004274 KB631763 ERL85933.1 KQ762329 OAD55966.1 GEZM01064783 JAV68642.1 UFQT01000257 SSX22451.1 KQ982254 KYQ58590.1 KK852528 KDR21990.1 CH954179 EDV54957.1 CH916367 EDW01282.1 GEBQ01031237 JAT08740.1 KQ977634 KYN01172.1 NEVH01005291 PNF38837.1 CH963849 EDW74078.1 GECU01017769 JAS89937.1 CH933808 EDW09896.1 ABLF02034549 ABLF02034555 ABLF02034557 ABLF02045036 CH940648 EDW61076.1 CP012524 ALC41188.1 LN830736 CFW94245.1 GECZ01004266 JAS65503.1 KQ435798 KOX73370.1 KQ434781 KZC04497.1 CM002911 KMY94656.1 SSX22450.1 EZ423158 ADD19434.1 CH480816 EDW48341.1 GFTR01004511 JAW11915.1 GFDF01002488 JAV11596.1 CM000071 EAL25767.2 GECL01000782 JAP05342.1 GEDC01027996 GEDC01022797 JAS09302.1 JAS14501.1 CH479183 EDW36224.1 GBBI01002698 JAC16014.1 GGFM01005340 MBW26091.1 GGFK01012274 MBW45595.1 GDHF01021405 JAI30909.1 GBGD01002441 JAC86448.1 CM000158 EDW91755.1 CH902619 EDV36991.1 ADMH02001863 ETN60799.1 CH477726 EAT36858.1 KA648077 AFP62706.1 AE013599 AF160934 LNIX01000025 OXA42603.1 OUUW01000001 SPP73268.1 GBXI01006909 JAD07383.1 GAPW01003375 JAC10223.1 GBHO01032889 GBHO01032883 GBHO01005527 GBHO01005524 GBRD01017016 JAG10715.1 JAG10721.1 JAG38077.1 JAG38080.1 JAG48811.1 GFDL01009282 JAV25763.1 HF549287 CCO62048.1 AAAB01008898 EAL40545.4 GEBF01000357 JAO03276.1 GACC01000052 JAA53755.1 AGTP01052648 AGTP01052649 AGTP01052650 AGTP01052651 AGTP01052652 AGTP01052653 AGTP01052654 AGTP01052655 AGTP01052656 AGTP01052657 AAKN02045239 AAKN02045240 AAKN02045241 CCAG010015795 AB077281 AB077282 AK153625 AK171340 AL596116 AL645687 AL645905 BC108351 GABZ01007299 JAA46226.1 ADFV01091842 ADFV01091843 ADFV01091844 ADFV01091845 ADFV01091846 ADFV01091847 ADFV01091848 ADFV01091849 ADFV01091850 ADFV01091851 NDHI03003438 PNJ50874.1 AANG04004184 AEYP01025685 AEYP01025686 AEYP01025687 AEYP01025688 AEYP01025689 AEYP01025690 AEYP01025691 AEYP01025692 AEYP01025693 AEYP01025694
Proteomes
UP000218220
UP000053240
UP000283053
UP000007151
UP000005204
UP000036403
+ More
UP000005203 UP000053097 UP000008237 UP000078540 UP000078492 UP000078541 UP000005205 UP000007266 UP000279307 UP000037069 UP000019118 UP000092462 UP000030742 UP000079169 UP000075809 UP000027135 UP000008711 UP000001070 UP000078542 UP000235965 UP000007798 UP000009192 UP000007819 UP000008792 UP000192223 UP000092553 UP000053105 UP000076502 UP000001292 UP000001819 UP000008744 UP000192221 UP000002282 UP000007801 UP000000673 UP000008820 UP000095301 UP000000803 UP000075884 UP000075885 UP000076407 UP000198287 UP000268350 UP000095300 UP000075881 UP000007062 UP000079721 UP000087266 UP000005215 UP000005447 UP000092444 UP000248480 UP000000589 UP000261540 UP000001073 UP000286641 UP000011712 UP000248482 UP000000715
UP000005203 UP000053097 UP000008237 UP000078540 UP000078492 UP000078541 UP000005205 UP000007266 UP000279307 UP000037069 UP000019118 UP000092462 UP000030742 UP000079169 UP000075809 UP000027135 UP000008711 UP000001070 UP000078542 UP000235965 UP000007798 UP000009192 UP000007819 UP000008792 UP000192223 UP000092553 UP000053105 UP000076502 UP000001292 UP000001819 UP000008744 UP000192221 UP000002282 UP000007801 UP000000673 UP000008820 UP000095301 UP000000803 UP000075884 UP000075885 UP000076407 UP000198287 UP000268350 UP000095300 UP000075881 UP000007062 UP000079721 UP000087266 UP000005215 UP000005447 UP000092444 UP000248480 UP000000589 UP000261540 UP000001073 UP000286641 UP000011712 UP000248482 UP000000715
Pfam
PF02121 IP_trans
Gene 3D
ProteinModelPortal
A0A2W1BZV5
A0A2A4IYV1
A0A2H1V1D9
A0A194QY14
A0A3S2M504
A0A212EQQ6
+ More
H9J752 A0A1E1VY14 A0A0C9QHL5 E9I8B4 V9IFY7 A0A0J7NV44 A0A087ZP11 A0A026VXE8 E2BF69 A0A195BJ38 A0A195DV06 A0A195F4N3 A0A158NJU3 D6WY44 A0A3L8DNS6 A0A0L0BNY7 J3JWK5 A0A1B0D981 U4U0X8 A0A1S3D367 A0A310SMK7 A0A1Y1L4M8 A0A336LZK0 A0A151XE44 A0A067RMZ4 B3NML8 B4J8L5 A0A1B6KBC2 A0A195CL38 A0A2J7RDC4 B4MPN9 A0A1B6ISQ8 B4KMX4 J9JMS4 B4LNC4 A0A1W4XBN7 A0A0M4EDS6 A0A0F7VK74 A0A1B6GT25 A0A0M8ZXX1 A0A154NY03 A0A0J9RFI1 A0A336LXC2 D3TP06 B4HMZ1 A0A224XHE9 A0A1L8DYT7 Q28Z48 A0A0V0GBI8 A0A1B6C760 B4GIA4 A0A023F4V1 A0A2M3ZC66 A0A2M4AXX7 A0A0K8UW46 A0A069DQW1 A0A1W4VCL4 B4P554 B3MIE3 W5JDA8 Q16R16 T1PGZ5 Q9U9P7 A0A182NSG3 A0A1S4FS70 A0A182PTC3 A0A182XFC5 A0A226DBQ2 A0A3B0JQ37 A0A0A1X9P8 A0A023ELS9 A0A1I8PZW0 A0A182JTZ5 A0A0A9YYI6 A0A1Q3FDX7 L8BSU6 Q5TSY2 A0A0P6JH43 A0A1S2ZTG3 K9IVU7 A0A1S3PJB2 I3N7H0 A0A286XCN4 A0A1B0FHV0 A0A2Y9DH67 Q8K4R4-2 K9IX44 A0A3B3RWG1 G1QL61 A0A2J8V029 A0A3Q7RY60 M3VY39 A0A2Y9KR92 M3YTB3
H9J752 A0A1E1VY14 A0A0C9QHL5 E9I8B4 V9IFY7 A0A0J7NV44 A0A087ZP11 A0A026VXE8 E2BF69 A0A195BJ38 A0A195DV06 A0A195F4N3 A0A158NJU3 D6WY44 A0A3L8DNS6 A0A0L0BNY7 J3JWK5 A0A1B0D981 U4U0X8 A0A1S3D367 A0A310SMK7 A0A1Y1L4M8 A0A336LZK0 A0A151XE44 A0A067RMZ4 B3NML8 B4J8L5 A0A1B6KBC2 A0A195CL38 A0A2J7RDC4 B4MPN9 A0A1B6ISQ8 B4KMX4 J9JMS4 B4LNC4 A0A1W4XBN7 A0A0M4EDS6 A0A0F7VK74 A0A1B6GT25 A0A0M8ZXX1 A0A154NY03 A0A0J9RFI1 A0A336LXC2 D3TP06 B4HMZ1 A0A224XHE9 A0A1L8DYT7 Q28Z48 A0A0V0GBI8 A0A1B6C760 B4GIA4 A0A023F4V1 A0A2M3ZC66 A0A2M4AXX7 A0A0K8UW46 A0A069DQW1 A0A1W4VCL4 B4P554 B3MIE3 W5JDA8 Q16R16 T1PGZ5 Q9U9P7 A0A182NSG3 A0A1S4FS70 A0A182PTC3 A0A182XFC5 A0A226DBQ2 A0A3B0JQ37 A0A0A1X9P8 A0A023ELS9 A0A1I8PZW0 A0A182JTZ5 A0A0A9YYI6 A0A1Q3FDX7 L8BSU6 Q5TSY2 A0A0P6JH43 A0A1S2ZTG3 K9IVU7 A0A1S3PJB2 I3N7H0 A0A286XCN4 A0A1B0FHV0 A0A2Y9DH67 Q8K4R4-2 K9IX44 A0A3B3RWG1 G1QL61 A0A2J8V029 A0A3Q7RY60 M3VY39 A0A2Y9KR92 M3YTB3
PDB
1T27
E-value=2.63757e-45,
Score=457
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
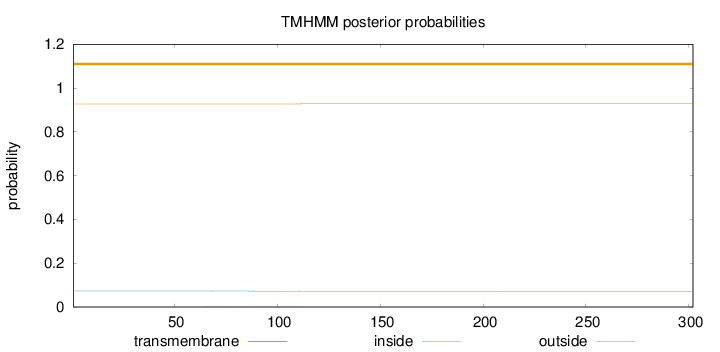
Length:
302
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01998
Exp number, first 60 AAs:
0.00035
Total prob of N-in:
0.07297
outside
1 - 302
Population Genetic Test Statistics
Pi
174.282872
Theta
189.187245
Tajima's D
-0.449118
CLR
23.980173
CSRT
0.253437328133593
Interpretation
Uncertain
