Gene
KWMTBOMO04537
Pre Gene Modal
BGIBMGA005463
Annotation
PREDICTED:_enteropeptidase_isoform_X1_[Bombyx_mori]
Full name
Enteropeptidase
Alternative Name
Enterokinase
Serine protease 7
Transmembrane protease serine 15
Serine protease 7
Transmembrane protease serine 15
Location in the cell
Extracellular Reliability : 3.299
Sequence
CDS
ATGTCGATACAGTCAGGCACCTTATCAAAAAACGTATCAGATAACGCCGATCGTAATAATTTTATAATGTATAAATTTTATGGTGCAAGTGTTAAAACTAGCGTAGTGCTGATTGTATTGTTTACTAAGTTGTGTGGTTTGACAGAATGTCGACGGGTTAAGAGAGTTGTTAACGGAAATGAAGTTCGATGCGGTACCGAACCTCGTGTGGGCTCCTTACATAGGAGAGACACACAGGAACATCTTTGTGGGGCAGTGGTTCTTTCTCGGAATTTCGCCATAACAGCAGCGCATTGTGTACATCTATCTGAAGACAATTATGTTCTCCAGCTTAATAATTACTGTATCAGAGATGGTGAAACTAGGCCAATAGCTAAAATTACGCAAATTATAACAAATGATCTGTATGATAGGGCATCAAAAGCGCACGACATTGCTTTGTTGAGAATTACACTAGATTTGAATGACGCGAACTCTGAATGGCTTAATGAATCAGTGCTGCCGAGCTCCTCATTTGGTGTGTCAGGTGAATGTACTATTTACGGCTACGGTTACAAGGATGTTAACACAATGAAGACATCGGATGTGTTGTCATCCGGTGAAGTTATAATAATTTCGCTGGACGAATGTACACAAAAATTGGGACCGTTCGTAGCGCCACAACCAGACAGCGGAATGATGTGCGCTATTGGTTATGGAGTAGATGCTTGTCAGGGTGATTCGGGTAGTCCCTTGATATGCGAAGGTTTAATACAAGGCATCAGTAGCTACGGAATGAGCTGCGGTGTATCGAATCTTCCCGGAATATACACCAGCATAGGTGCTCACCTGGACTTCATACAAAACGCAATTCAAACACAGAACCAAATAAAAACCAATGCTTCTGCGAATATTGAATAA
Protein
MSIQSGTLSKNVSDNADRNNFIMYKFYGASVKTSVVLIVLFTKLCGLTECRRVKRVVNGNEVRCGTEPRVGSLHRRDTQEHLCGAVVLSRNFAITAAHCVHLSEDNYVLQLNNYCIRDGETRPIAKITQIITNDLYDRASKAHDIALLRITLDLNDANSEWLNESVLPSSSFGVSGECTIYGYGYKDVNTMKTSDVLSSGEVIIISLDECTQKLGPFVAPQPDSGMMCAIGYGVDACQGDSGSPLICEGLIQGISSYGMSCGVSNLPGIYTSIGAHLDFIQNAIQTQNQIKTNASANIE
Summary
Description
Responsible for initiating activation of pancreatic proteolytic proenzymes (trypsin, chymotrypsin and carboxypeptidase A). It catalyzes the conversion of trypsinogen to trypsin which in turn activates other proenzymes including chymotrypsinogen, procarboxypeptidases, and proelastases.
Responsible for initiating activation of pancreatic proteolytic proenzymes (trypsin, chymotrypsin and carboxypeptidase A). It catalyzes the conversion of trypsinogen to trypsin which in turn activates other proenzymes including chymotrypsinogen, procarboxypeptidases, and proelastases (By similarity).
Responsible for initiating activation of pancreatic proteolytic proenzymes (trypsin, chymotrypsin and carboxypeptidase A). It catalyzes the conversion of trypsinogen to trypsin which in turn activates other proenzymes including chymotrypsinogen, procarboxypeptidases, and proelastases (By similarity).
Catalytic Activity
Activation of trypsinogen by selective cleavage of 6-Lys-|-Ile-7 bond.
Subunit
Heterodimer of a catalytic (light) chain and a multidomain (heavy) chain linked by a disulfide bond.
Similarity
Belongs to the peptidase S1 family.
Keywords
Complete proteome
Disulfide bond
Glycoprotein
Hydrolase
Membrane
Protease
Reference proteome
Repeat
Serine protease
Signal-anchor
Transmembrane
Transmembrane helix
Zymogen
Feature
chain Enteropeptidase non-catalytic heavy chain
Uniprot
A0A2W1BZT5
A0A2A4IX92
A0A2H1V1D1
A0A194QTZ4
A0A194PM44
E9GB32
+ More
A0A0P5F3R6 A0A0P4XVX6 A0A0P6I447 A0A3B3C1C2 A0A0P6CUK1 A0A0P6GIJ8 A0A0P5DIE6 A0A146UZ51 A0A3B3C1X5 A0A3B4AC48 A0A0N8DHB8 E9Q6Y6 P97435 H3DEU3 A0A146UXL9 A0A1L8FLU3 A0A146UXA2 A0A3Q2Q818 A0A146UXJ5 A0A2M3ZE40 A0A3B5LVW8 A0A146UXG2 A0A1A8AXS9 A0A3Q1FHB2 A0A3B4AC44 A0A1A8S8U1 A0A3Q3X1C1 A0A1W4ZC14 M4A4V5 Q8CAN9 H3D589 A0A182FII0 A0A3B4AAH6 A0A2M4BVE0 A0A3Q3M963 A0A2M3Z7J6 R0JV69 A0A3S2P3L8 A0A3P9P1R4 A0A3S2MRB3 A0A3B5LQZ5 A0A3Q1CVP0 A0A3B3BF20 A0A3P8SG71 A0A3Q3MTZ1 A0A2M4BV71 U5EQV5 A0A3B3BG67 A0A3B3IM09 A0A3P9JRW9 A4UWM5 A0A3P9JR68 A4UWM6 A0A3P9KGZ9 A0A3B4UG61 A0A3P9HXI9 A0A1U8BJU8 A0A3B4WG00 I3JYM8 A0A3B5BL60 A0A3Q3CW68 A0A3B3XES2 A0A3Q0RDT6 A0A2U9B4D3 A0A3P9M9M9 A0A0P5ZIR4 A0A3P9KWP6 A0A3Q0SR17 A0A099ZE42 R4QR01 W5JKI4 A0A3B4BRU8 A0A3B3TT46 A0A3B4WN70 E6Y432 A0A3P9AGM1 Q6B4R4 W5Q241 A0A2U9B712 A0A146Y9Y3 A0A3Q3IQH3 A0A147AUU9 A0A1S3KCW5 G3NFA1 A0A3B4GGL4 A0A3Q2ZWF1 A0A060XBB7 A0A3P9BHV1 A0A3Q4N5U5 A0A146YBK3 A0A1S3NSU4
A0A0P5F3R6 A0A0P4XVX6 A0A0P6I447 A0A3B3C1C2 A0A0P6CUK1 A0A0P6GIJ8 A0A0P5DIE6 A0A146UZ51 A0A3B3C1X5 A0A3B4AC48 A0A0N8DHB8 E9Q6Y6 P97435 H3DEU3 A0A146UXL9 A0A1L8FLU3 A0A146UXA2 A0A3Q2Q818 A0A146UXJ5 A0A2M3ZE40 A0A3B5LVW8 A0A146UXG2 A0A1A8AXS9 A0A3Q1FHB2 A0A3B4AC44 A0A1A8S8U1 A0A3Q3X1C1 A0A1W4ZC14 M4A4V5 Q8CAN9 H3D589 A0A182FII0 A0A3B4AAH6 A0A2M4BVE0 A0A3Q3M963 A0A2M3Z7J6 R0JV69 A0A3S2P3L8 A0A3P9P1R4 A0A3S2MRB3 A0A3B5LQZ5 A0A3Q1CVP0 A0A3B3BF20 A0A3P8SG71 A0A3Q3MTZ1 A0A2M4BV71 U5EQV5 A0A3B3BG67 A0A3B3IM09 A0A3P9JRW9 A4UWM5 A0A3P9JR68 A4UWM6 A0A3P9KGZ9 A0A3B4UG61 A0A3P9HXI9 A0A1U8BJU8 A0A3B4WG00 I3JYM8 A0A3B5BL60 A0A3Q3CW68 A0A3B3XES2 A0A3Q0RDT6 A0A2U9B4D3 A0A3P9M9M9 A0A0P5ZIR4 A0A3P9KWP6 A0A3Q0SR17 A0A099ZE42 R4QR01 W5JKI4 A0A3B4BRU8 A0A3B3TT46 A0A3B4WN70 E6Y432 A0A3P9AGM1 Q6B4R4 W5Q241 A0A2U9B712 A0A146Y9Y3 A0A3Q3IQH3 A0A147AUU9 A0A1S3KCW5 G3NFA1 A0A3B4GGL4 A0A3Q2ZWF1 A0A060XBB7 A0A3P9BHV1 A0A3Q4N5U5 A0A146YBK3 A0A1S3NSU4
EC Number
3.4.21.9
Pubmed
EMBL
KZ149904
PZC78370.1
NWSH01005209
PCG64351.1
ODYU01000234
SOQ34660.1
+ More
KQ461154 KPJ08465.1 KQ459599 KPI94397.1 GL732537 EFX83438.1 GDIP01151859 JAJ71543.1 GDIP01237251 JAI86150.1 GDIQ01026413 JAN68324.1 GDIP01078763 GDIQ01090111 JAM24952.1 JAN04626.1 GDIQ01034232 JAN60505.1 GDIP01156858 JAJ66544.1 GCES01076476 JAR09847.1 GDIP01033703 JAM70012.1 AC112257 AC161815 U73378 BC117917 BC117918 GCES01076472 JAR09851.1 CM004478 OCT72566.1 GCES01076473 JAR09850.1 GCES01076470 JAR09853.1 GGFM01006055 MBW26806.1 GCES01076471 JAR09852.1 HADY01021497 SBP59982.1 HAEH01022436 SBS13959.1 AK038356 BAC29973.1 GGFJ01007842 MBW56983.1 GGFM01003733 MBW24484.1 KB743121 EOB01127.1 CM012450 RVE63431.1 CM012449 RVE64986.1 GGFJ01007826 MBW56967.1 GANO01004076 JAB55795.1 AB272104 BAF57203.1 AB272105 BAF57204.1 AERX01036422 CP026245 AWO98823.1 GDIP01042976 JAM60739.1 KL893329 KGL80679.1 KC756844 AGL73301.1 ADMH02001179 ETN63823.1 EU375507 ACB10253.1 AY682203 AAT84164.1 AMGL01006515 AMGL01006516 AMGL01006517 AMGL01006518 AMGL01006519 AMGL01006520 AMGL01006521 AMGL01006522 CP026246 AWO99724.1 GCES01035362 JAR50961.1 GCES01004398 JAR81925.1 FR905186 CDQ76928.1 GCES01035358 JAR50965.1
KQ461154 KPJ08465.1 KQ459599 KPI94397.1 GL732537 EFX83438.1 GDIP01151859 JAJ71543.1 GDIP01237251 JAI86150.1 GDIQ01026413 JAN68324.1 GDIP01078763 GDIQ01090111 JAM24952.1 JAN04626.1 GDIQ01034232 JAN60505.1 GDIP01156858 JAJ66544.1 GCES01076476 JAR09847.1 GDIP01033703 JAM70012.1 AC112257 AC161815 U73378 BC117917 BC117918 GCES01076472 JAR09851.1 CM004478 OCT72566.1 GCES01076473 JAR09850.1 GCES01076470 JAR09853.1 GGFM01006055 MBW26806.1 GCES01076471 JAR09852.1 HADY01021497 SBP59982.1 HAEH01022436 SBS13959.1 AK038356 BAC29973.1 GGFJ01007842 MBW56983.1 GGFM01003733 MBW24484.1 KB743121 EOB01127.1 CM012450 RVE63431.1 CM012449 RVE64986.1 GGFJ01007826 MBW56967.1 GANO01004076 JAB55795.1 AB272104 BAF57203.1 AB272105 BAF57204.1 AERX01036422 CP026245 AWO98823.1 GDIP01042976 JAM60739.1 KL893329 KGL80679.1 KC756844 AGL73301.1 ADMH02001179 ETN63823.1 EU375507 ACB10253.1 AY682203 AAT84164.1 AMGL01006515 AMGL01006516 AMGL01006517 AMGL01006518 AMGL01006519 AMGL01006520 AMGL01006521 AMGL01006522 CP026246 AWO99724.1 GCES01035362 JAR50961.1 GCES01004398 JAR81925.1 FR905186 CDQ76928.1 GCES01035358 JAR50965.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000000305
UP000261560
UP000261520
+ More
UP000000589 UP000007303 UP000186698 UP000265000 UP000261380 UP000257200 UP000261620 UP000192224 UP000002852 UP000069272 UP000261640 UP000242638 UP000257160 UP000265080 UP000001038 UP000265200 UP000265180 UP000261420 UP000189706 UP000261360 UP000005207 UP000261400 UP000264840 UP000261480 UP000261340 UP000246464 UP000053641 UP000000673 UP000261440 UP000261500 UP000265140 UP000002356 UP000261600 UP000085678 UP000007635 UP000261460 UP000264800 UP000193380 UP000265160 UP000261580 UP000087266
UP000000589 UP000007303 UP000186698 UP000265000 UP000261380 UP000257200 UP000261620 UP000192224 UP000002852 UP000069272 UP000261640 UP000242638 UP000257160 UP000265080 UP000001038 UP000265200 UP000265180 UP000261420 UP000189706 UP000261360 UP000005207 UP000261400 UP000264840 UP000261480 UP000261340 UP000246464 UP000053641 UP000000673 UP000261440 UP000261500 UP000265140 UP000002356 UP000261600 UP000085678 UP000007635 UP000261460 UP000264800 UP000193380 UP000265160 UP000261580 UP000087266
Pfam
Interpro
IPR033116
TRYPSIN_SER
+ More
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR001190 SRCR
IPR036772 SRCR-like_dom_sf
IPR017448 SRCR-like_dom
IPR002172 LDrepeatLR_classA_rpt
IPR000859 CUB_dom
IPR035914 Sperma_CUB_dom_sf
IPR036055 LDL_receptor-like_sf
IPR013320 ConA-like_dom_sf
IPR036364 SEA_dom_sf
IPR000082 SEA_dom
IPR011163 Pept_S1A_enterop
IPR000998 MAM_dom
IPR023415 LDLR_class-A_CS
IPR012224 Pept_S1A_FX
IPR015352 Hepsin-SRCR_dom
IPR017949 Thaumatin_CS
IPR011333 SKP1/BTB/POZ_sf
IPR036236 Znf_C2H2_sf
IPR000210 BTB/POZ_dom
IPR013087 Znf_C2H2_type
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR001190 SRCR
IPR036772 SRCR-like_dom_sf
IPR017448 SRCR-like_dom
IPR002172 LDrepeatLR_classA_rpt
IPR000859 CUB_dom
IPR035914 Sperma_CUB_dom_sf
IPR036055 LDL_receptor-like_sf
IPR013320 ConA-like_dom_sf
IPR036364 SEA_dom_sf
IPR000082 SEA_dom
IPR011163 Pept_S1A_enterop
IPR000998 MAM_dom
IPR023415 LDLR_class-A_CS
IPR012224 Pept_S1A_FX
IPR015352 Hepsin-SRCR_dom
IPR017949 Thaumatin_CS
IPR011333 SKP1/BTB/POZ_sf
IPR036236 Znf_C2H2_sf
IPR000210 BTB/POZ_dom
IPR013087 Znf_C2H2_type
SUPFAM
ProteinModelPortal
A0A2W1BZT5
A0A2A4IX92
A0A2H1V1D1
A0A194QTZ4
A0A194PM44
E9GB32
+ More
A0A0P5F3R6 A0A0P4XVX6 A0A0P6I447 A0A3B3C1C2 A0A0P6CUK1 A0A0P6GIJ8 A0A0P5DIE6 A0A146UZ51 A0A3B3C1X5 A0A3B4AC48 A0A0N8DHB8 E9Q6Y6 P97435 H3DEU3 A0A146UXL9 A0A1L8FLU3 A0A146UXA2 A0A3Q2Q818 A0A146UXJ5 A0A2M3ZE40 A0A3B5LVW8 A0A146UXG2 A0A1A8AXS9 A0A3Q1FHB2 A0A3B4AC44 A0A1A8S8U1 A0A3Q3X1C1 A0A1W4ZC14 M4A4V5 Q8CAN9 H3D589 A0A182FII0 A0A3B4AAH6 A0A2M4BVE0 A0A3Q3M963 A0A2M3Z7J6 R0JV69 A0A3S2P3L8 A0A3P9P1R4 A0A3S2MRB3 A0A3B5LQZ5 A0A3Q1CVP0 A0A3B3BF20 A0A3P8SG71 A0A3Q3MTZ1 A0A2M4BV71 U5EQV5 A0A3B3BG67 A0A3B3IM09 A0A3P9JRW9 A4UWM5 A0A3P9JR68 A4UWM6 A0A3P9KGZ9 A0A3B4UG61 A0A3P9HXI9 A0A1U8BJU8 A0A3B4WG00 I3JYM8 A0A3B5BL60 A0A3Q3CW68 A0A3B3XES2 A0A3Q0RDT6 A0A2U9B4D3 A0A3P9M9M9 A0A0P5ZIR4 A0A3P9KWP6 A0A3Q0SR17 A0A099ZE42 R4QR01 W5JKI4 A0A3B4BRU8 A0A3B3TT46 A0A3B4WN70 E6Y432 A0A3P9AGM1 Q6B4R4 W5Q241 A0A2U9B712 A0A146Y9Y3 A0A3Q3IQH3 A0A147AUU9 A0A1S3KCW5 G3NFA1 A0A3B4GGL4 A0A3Q2ZWF1 A0A060XBB7 A0A3P9BHV1 A0A3Q4N5U5 A0A146YBK3 A0A1S3NSU4
A0A0P5F3R6 A0A0P4XVX6 A0A0P6I447 A0A3B3C1C2 A0A0P6CUK1 A0A0P6GIJ8 A0A0P5DIE6 A0A146UZ51 A0A3B3C1X5 A0A3B4AC48 A0A0N8DHB8 E9Q6Y6 P97435 H3DEU3 A0A146UXL9 A0A1L8FLU3 A0A146UXA2 A0A3Q2Q818 A0A146UXJ5 A0A2M3ZE40 A0A3B5LVW8 A0A146UXG2 A0A1A8AXS9 A0A3Q1FHB2 A0A3B4AC44 A0A1A8S8U1 A0A3Q3X1C1 A0A1W4ZC14 M4A4V5 Q8CAN9 H3D589 A0A182FII0 A0A3B4AAH6 A0A2M4BVE0 A0A3Q3M963 A0A2M3Z7J6 R0JV69 A0A3S2P3L8 A0A3P9P1R4 A0A3S2MRB3 A0A3B5LQZ5 A0A3Q1CVP0 A0A3B3BF20 A0A3P8SG71 A0A3Q3MTZ1 A0A2M4BV71 U5EQV5 A0A3B3BG67 A0A3B3IM09 A0A3P9JRW9 A4UWM5 A0A3P9JR68 A4UWM6 A0A3P9KGZ9 A0A3B4UG61 A0A3P9HXI9 A0A1U8BJU8 A0A3B4WG00 I3JYM8 A0A3B5BL60 A0A3Q3CW68 A0A3B3XES2 A0A3Q0RDT6 A0A2U9B4D3 A0A3P9M9M9 A0A0P5ZIR4 A0A3P9KWP6 A0A3Q0SR17 A0A099ZE42 R4QR01 W5JKI4 A0A3B4BRU8 A0A3B3TT46 A0A3B4WN70 E6Y432 A0A3P9AGM1 Q6B4R4 W5Q241 A0A2U9B712 A0A146Y9Y3 A0A3Q3IQH3 A0A147AUU9 A0A1S3KCW5 G3NFA1 A0A3B4GGL4 A0A3Q2ZWF1 A0A060XBB7 A0A3P9BHV1 A0A3Q4N5U5 A0A146YBK3 A0A1S3NSU4
PDB
3W94
E-value=3.11117e-26,
Score=293
Ontologies
GO
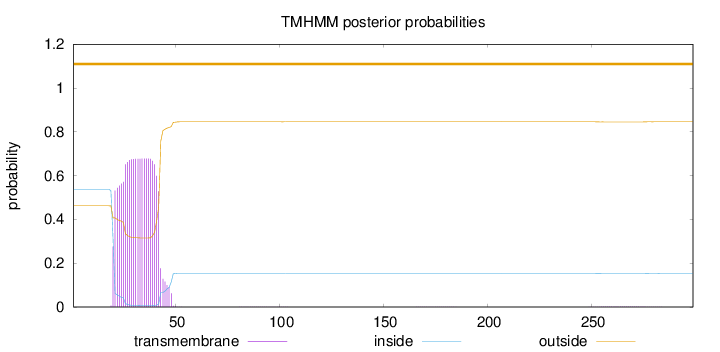
Topology
Subcellular location
Membrane
Length:
299
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
14.99718
Exp number, first 60 AAs:
14.92727
Total prob of N-in:
0.53780
POSSIBLE N-term signal
sequence
outside
1 - 299
Population Genetic Test Statistics
Pi
234.552375
Theta
172.518847
Tajima's D
1.313062
CLR
0.163161
CSRT
0.753212339383031
Interpretation
Uncertain
