Pre Gene Modal
BGIBMGA005462
Annotation
PREDICTED:_NADH_dehydrogenase_[ubiquinone]_1_alpha_subcomplex_subunit_6-like_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 3.323
Sequence
CDS
ATGTCTGCAAGGCAGGCAATAAAAGTTGGTACAAAAACCGTAAAACCAGTACTATCGTCGTCTCATGCTGAAGCTCGGAATAGAGTTCTGAGTCTTTACAAAGCATGGTATCGTCAGATCCCCTATATAGTGAAGGATTATGACATTCCCAAGTCAGAAGCACAATGTCGAGAGAAATTGAAAGAACTCTTCATTAAGAACAAACATGTGACTGATATTCGAGTCATTGATATGCTGGTTATTAAGGGTCAAATGGAGTTGAAAGAGTCGGTGAACATATGGAAACAAAAGGGGCACATTATGGCCTACTTCAAACCAACAGAAGAACCTAAACCAAAAAATTTCCTTTCGAAATTCTTTTCTGGAAACGAATGA
Protein
MSARQAIKVGTKTVKPVLSSSHAEARNRVLSLYKAWYRQIPYIVKDYDIPKSEAQCREKLKELFIKNKHVTDIRVIDMLVIKGQMELKESVNIWKQKGHIMAYFKPTEEPKPKNFLSKFFSGNE
Summary
Similarity
Belongs to the complex I LYR family.
Belongs to the tetraspanin (TM4SF) family.
Belongs to the tetraspanin (TM4SF) family.
Uniprot
H9J7H0
S4P9V1
A0A212EJF9
I4DK69
A0A194PLT7
A0A2W1BYS3
+ More
A0A2H1VYZ0 A0A194QSJ9 A0A1E1W373 A0A182FPE2 A0A1B0CJY0 A0A2M3Z512 A0A2P8Z6T4 A0A1Q3FL65 A0A0L7LUQ5 A0A336KWV3 A0A182VXQ7 A0A182RJ79 A0A336M644 A0A182GAU6 A0A182XXA2 A0A2M4C3K4 T1DKV1 A0A023EDZ8 A0A182QQJ1 Q17Q29 A0A182PBG8 A0A182KK73 A0A182XKF2 A0A182KFR6 B0WQF7 W5JGV3 A0A1L8E0P9 A0A182NLA0 A0A2M4AEF7 Q7QM15 A0A182IKX4 Q7Q9C1 A0A182HJJ2 A0A182UXX9 A0A2J7QDN1 A0A182M072 B4MRY0 Q28ZU9 B4LJC1 A0A1B0CZR5 B4JVZ0 A0A1A9URC8 A0A1A9Z2G6 B4P7M4 B3NN29 A0A3B0JNX9 T1PFB1 A0A1B0FK01 D1FQ06 A0A1W4WSQ1 A0A1L8EER1 A0A1A9WWU7 A0A1I8QAR8 A0A1A9YL53 A0A069DMX4 B3MCS3 B4NSC9 Q7JZK1 A0A1W4VCJ1 A0A0A9WBQ8 B4HN24 A0A0M4ETG2 N6U0D3 B4GGR1 B4KTT1 A0A0N7Z9G2 R4FPF4 U4U7N1 A0A161MLJ2 W8C2I8 A0A1B0C5E2 A0A0P4WG23 A0A2R7WG33 A0A0P5FVM7 A0A162QNW1 A0A0V0G6U0 A0A0L0C5I4 A0A0N8BP27 A0A0P6F0P7 A0A023F825 A0A0K8V405 A0A0P5F9X0 A0A0P5MI13 A0A0P5NJT7 A0A226EDV4 A0A034VMX7 D7EHX8 A0A3R7N0B5 A0A0P6JXI5 A0A0P5D5G1 A0A0P5KCS9 A0A0P5EQ33 A0A0P5M2C4 A0A0P5DPX8
A0A2H1VYZ0 A0A194QSJ9 A0A1E1W373 A0A182FPE2 A0A1B0CJY0 A0A2M3Z512 A0A2P8Z6T4 A0A1Q3FL65 A0A0L7LUQ5 A0A336KWV3 A0A182VXQ7 A0A182RJ79 A0A336M644 A0A182GAU6 A0A182XXA2 A0A2M4C3K4 T1DKV1 A0A023EDZ8 A0A182QQJ1 Q17Q29 A0A182PBG8 A0A182KK73 A0A182XKF2 A0A182KFR6 B0WQF7 W5JGV3 A0A1L8E0P9 A0A182NLA0 A0A2M4AEF7 Q7QM15 A0A182IKX4 Q7Q9C1 A0A182HJJ2 A0A182UXX9 A0A2J7QDN1 A0A182M072 B4MRY0 Q28ZU9 B4LJC1 A0A1B0CZR5 B4JVZ0 A0A1A9URC8 A0A1A9Z2G6 B4P7M4 B3NN29 A0A3B0JNX9 T1PFB1 A0A1B0FK01 D1FQ06 A0A1W4WSQ1 A0A1L8EER1 A0A1A9WWU7 A0A1I8QAR8 A0A1A9YL53 A0A069DMX4 B3MCS3 B4NSC9 Q7JZK1 A0A1W4VCJ1 A0A0A9WBQ8 B4HN24 A0A0M4ETG2 N6U0D3 B4GGR1 B4KTT1 A0A0N7Z9G2 R4FPF4 U4U7N1 A0A161MLJ2 W8C2I8 A0A1B0C5E2 A0A0P4WG23 A0A2R7WG33 A0A0P5FVM7 A0A162QNW1 A0A0V0G6U0 A0A0L0C5I4 A0A0N8BP27 A0A0P6F0P7 A0A023F825 A0A0K8V405 A0A0P5F9X0 A0A0P5MI13 A0A0P5NJT7 A0A226EDV4 A0A034VMX7 D7EHX8 A0A3R7N0B5 A0A0P6JXI5 A0A0P5D5G1 A0A0P5KCS9 A0A0P5EQ33 A0A0P5M2C4 A0A0P5DPX8
Pubmed
19121390
23622113
22118469
22651552
26354079
28756777
+ More
29403074 26227816 26483478 25244985 24945155 17510324 26999592 20966253 20920257 23761445 9087549 12364791 14747013 17210077 17994087 15632085 17550304 25315136 26334808 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 26823975 23537049 27129103 24495485 26108605 25474469 25348373 18362917 19820115
29403074 26227816 26483478 25244985 24945155 17510324 26999592 20966253 20920257 23761445 9087549 12364791 14747013 17210077 17994087 15632085 17550304 25315136 26334808 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 26823975 23537049 27129103 24495485 26108605 25474469 25348373 18362917 19820115
EMBL
BABH01013834
GAIX01003559
JAA89001.1
AGBW02014484
OWR41600.1
AK401687
+ More
BAM18309.1 KQ459599 KPI94401.1 KZ149904 PZC78387.1 ODYU01005326 SOQ46060.1 KQ461154 KPJ08462.1 GDQN01009629 JAT81425.1 AJWK01015524 AJWK01015525 GGFM01002858 MBW23609.1 PYGN01000170 PSN52207.1 GFDL01006695 JAV28350.1 JTDY01000051 KOB79165.1 UFQS01000611 UFQT01000611 SSX05370.1 SSX25731.1 SSX05365.1 SSX25726.1 JXUM01008769 JXUM01008770 KQ560268 KXJ83442.1 GGFJ01010742 MBW59883.1 GAMD01000766 JAB00825.1 GAPW01006061 JAC07537.1 AXCN02000114 CH477187 GDUN01001025 EAT48887.1 JAN94894.1 DS232039 EDS32828.1 ADMH02001538 ETN62135.1 GFDF01001803 JAV12281.1 GGFK01005829 MBW39150.1 AAAB01005603 EAA02579.3 AAAB01008904 EAA09633.5 APCN01002170 NEVH01015817 PNF26688.1 AXCM01015740 CH963850 EDW74869.1 CM000071 EAL25513.1 CH940648 EDW60501.1 AJVK01009695 CH916375 EDV98128.1 CM000158 EDW90059.1 CH954179 EDV56549.1 OUUW01000001 SPP75036.1 KA647431 AFP62060.1 CCAG010020733 EZ419906 ACZ28261.1 GFDG01001633 JAV17166.1 GBGD01003486 JAC85403.1 CH902619 EDV37325.1 CH982158 CH982159 CM002911 EDX15507.1 EDX15511.1 KMY92889.1 AE013599 AY071209 AAF58729.1 AAL48831.1 AGB93410.1 GBHO01038768 GBRD01004963 GDHC01004245 JAG04836.1 JAG60858.1 JAQ14384.1 CH480816 EDW47328.1 CP012524 ALC40532.1 APGK01044294 APGK01044295 KB741022 ENN74995.1 CH479183 EDW35681.1 CH933808 EDW10657.1 GDKW01000635 JAI55960.1 ACPB03020118 GAHY01000909 JAA76601.1 KB631843 ERL86631.1 GEMB01002303 JAS00878.1 GAMC01006524 JAC00032.1 JXJN01025945 GDRN01027129 JAI67659.1 KK854775 PTY18603.1 GDIQ01251117 JAK00608.1 LRGB01000311 KZS19835.1 GECL01002451 JAP03673.1 JRES01000890 KNC27545.1 GDIQ01158843 JAK92882.1 GDIQ01055384 JAN39353.1 GBBI01001328 JAC17384.1 GDHF01019019 JAI33295.1 GDIQ01261207 JAJ90517.1 GDIQ01158844 JAK92881.1 GDIQ01141202 JAL10524.1 LNIX01000004 OXA55589.1 GAKP01015138 GAKP01015136 GAKP01015135 JAC43814.1 KQ971323 KYB28574.1 QCYY01001978 ROT73863.1 GDIQ01004446 JAN90291.1 GDIP01160959 JAJ62443.1 GDIQ01187533 JAK64192.1 GDIP01253293 GDIQ01268918 JAI70108.1 JAJ82806.1 GDIQ01170535 JAK81190.1 GDIP01156612 JAJ66790.1
BAM18309.1 KQ459599 KPI94401.1 KZ149904 PZC78387.1 ODYU01005326 SOQ46060.1 KQ461154 KPJ08462.1 GDQN01009629 JAT81425.1 AJWK01015524 AJWK01015525 GGFM01002858 MBW23609.1 PYGN01000170 PSN52207.1 GFDL01006695 JAV28350.1 JTDY01000051 KOB79165.1 UFQS01000611 UFQT01000611 SSX05370.1 SSX25731.1 SSX05365.1 SSX25726.1 JXUM01008769 JXUM01008770 KQ560268 KXJ83442.1 GGFJ01010742 MBW59883.1 GAMD01000766 JAB00825.1 GAPW01006061 JAC07537.1 AXCN02000114 CH477187 GDUN01001025 EAT48887.1 JAN94894.1 DS232039 EDS32828.1 ADMH02001538 ETN62135.1 GFDF01001803 JAV12281.1 GGFK01005829 MBW39150.1 AAAB01005603 EAA02579.3 AAAB01008904 EAA09633.5 APCN01002170 NEVH01015817 PNF26688.1 AXCM01015740 CH963850 EDW74869.1 CM000071 EAL25513.1 CH940648 EDW60501.1 AJVK01009695 CH916375 EDV98128.1 CM000158 EDW90059.1 CH954179 EDV56549.1 OUUW01000001 SPP75036.1 KA647431 AFP62060.1 CCAG010020733 EZ419906 ACZ28261.1 GFDG01001633 JAV17166.1 GBGD01003486 JAC85403.1 CH902619 EDV37325.1 CH982158 CH982159 CM002911 EDX15507.1 EDX15511.1 KMY92889.1 AE013599 AY071209 AAF58729.1 AAL48831.1 AGB93410.1 GBHO01038768 GBRD01004963 GDHC01004245 JAG04836.1 JAG60858.1 JAQ14384.1 CH480816 EDW47328.1 CP012524 ALC40532.1 APGK01044294 APGK01044295 KB741022 ENN74995.1 CH479183 EDW35681.1 CH933808 EDW10657.1 GDKW01000635 JAI55960.1 ACPB03020118 GAHY01000909 JAA76601.1 KB631843 ERL86631.1 GEMB01002303 JAS00878.1 GAMC01006524 JAC00032.1 JXJN01025945 GDRN01027129 JAI67659.1 KK854775 PTY18603.1 GDIQ01251117 JAK00608.1 LRGB01000311 KZS19835.1 GECL01002451 JAP03673.1 JRES01000890 KNC27545.1 GDIQ01158843 JAK92882.1 GDIQ01055384 JAN39353.1 GBBI01001328 JAC17384.1 GDHF01019019 JAI33295.1 GDIQ01261207 JAJ90517.1 GDIQ01158844 JAK92881.1 GDIQ01141202 JAL10524.1 LNIX01000004 OXA55589.1 GAKP01015138 GAKP01015136 GAKP01015135 JAC43814.1 KQ971323 KYB28574.1 QCYY01001978 ROT73863.1 GDIQ01004446 JAN90291.1 GDIP01160959 JAJ62443.1 GDIQ01187533 JAK64192.1 GDIP01253293 GDIQ01268918 JAI70108.1 JAJ82806.1 GDIQ01170535 JAK81190.1 GDIP01156612 JAJ66790.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000069272
UP000092461
+ More
UP000245037 UP000037510 UP000075920 UP000075900 UP000069940 UP000249989 UP000076408 UP000075886 UP000008820 UP000075885 UP000075882 UP000076407 UP000075881 UP000002320 UP000000673 UP000075884 UP000007062 UP000075880 UP000075840 UP000075903 UP000235965 UP000075883 UP000007798 UP000001819 UP000008792 UP000092462 UP000001070 UP000078200 UP000092445 UP000002282 UP000008711 UP000268350 UP000095301 UP000092444 UP000192223 UP000091820 UP000095300 UP000092443 UP000007801 UP000000304 UP000000803 UP000192221 UP000001292 UP000092553 UP000019118 UP000008744 UP000009192 UP000015103 UP000030742 UP000092460 UP000076858 UP000037069 UP000198287 UP000007266 UP000283509
UP000245037 UP000037510 UP000075920 UP000075900 UP000069940 UP000249989 UP000076408 UP000075886 UP000008820 UP000075885 UP000075882 UP000076407 UP000075881 UP000002320 UP000000673 UP000075884 UP000007062 UP000075880 UP000075840 UP000075903 UP000235965 UP000075883 UP000007798 UP000001819 UP000008792 UP000092462 UP000001070 UP000078200 UP000092445 UP000002282 UP000008711 UP000268350 UP000095301 UP000092444 UP000192223 UP000091820 UP000095300 UP000092443 UP000007801 UP000000304 UP000000803 UP000192221 UP000001292 UP000092553 UP000019118 UP000008744 UP000009192 UP000015103 UP000030742 UP000092460 UP000076858 UP000037069 UP000198287 UP000007266 UP000283509
Interpro
SUPFAM
SSF48652
SSF48652
Gene 3D
ProteinModelPortal
H9J7H0
S4P9V1
A0A212EJF9
I4DK69
A0A194PLT7
A0A2W1BYS3
+ More
A0A2H1VYZ0 A0A194QSJ9 A0A1E1W373 A0A182FPE2 A0A1B0CJY0 A0A2M3Z512 A0A2P8Z6T4 A0A1Q3FL65 A0A0L7LUQ5 A0A336KWV3 A0A182VXQ7 A0A182RJ79 A0A336M644 A0A182GAU6 A0A182XXA2 A0A2M4C3K4 T1DKV1 A0A023EDZ8 A0A182QQJ1 Q17Q29 A0A182PBG8 A0A182KK73 A0A182XKF2 A0A182KFR6 B0WQF7 W5JGV3 A0A1L8E0P9 A0A182NLA0 A0A2M4AEF7 Q7QM15 A0A182IKX4 Q7Q9C1 A0A182HJJ2 A0A182UXX9 A0A2J7QDN1 A0A182M072 B4MRY0 Q28ZU9 B4LJC1 A0A1B0CZR5 B4JVZ0 A0A1A9URC8 A0A1A9Z2G6 B4P7M4 B3NN29 A0A3B0JNX9 T1PFB1 A0A1B0FK01 D1FQ06 A0A1W4WSQ1 A0A1L8EER1 A0A1A9WWU7 A0A1I8QAR8 A0A1A9YL53 A0A069DMX4 B3MCS3 B4NSC9 Q7JZK1 A0A1W4VCJ1 A0A0A9WBQ8 B4HN24 A0A0M4ETG2 N6U0D3 B4GGR1 B4KTT1 A0A0N7Z9G2 R4FPF4 U4U7N1 A0A161MLJ2 W8C2I8 A0A1B0C5E2 A0A0P4WG23 A0A2R7WG33 A0A0P5FVM7 A0A162QNW1 A0A0V0G6U0 A0A0L0C5I4 A0A0N8BP27 A0A0P6F0P7 A0A023F825 A0A0K8V405 A0A0P5F9X0 A0A0P5MI13 A0A0P5NJT7 A0A226EDV4 A0A034VMX7 D7EHX8 A0A3R7N0B5 A0A0P6JXI5 A0A0P5D5G1 A0A0P5KCS9 A0A0P5EQ33 A0A0P5M2C4 A0A0P5DPX8
A0A2H1VYZ0 A0A194QSJ9 A0A1E1W373 A0A182FPE2 A0A1B0CJY0 A0A2M3Z512 A0A2P8Z6T4 A0A1Q3FL65 A0A0L7LUQ5 A0A336KWV3 A0A182VXQ7 A0A182RJ79 A0A336M644 A0A182GAU6 A0A182XXA2 A0A2M4C3K4 T1DKV1 A0A023EDZ8 A0A182QQJ1 Q17Q29 A0A182PBG8 A0A182KK73 A0A182XKF2 A0A182KFR6 B0WQF7 W5JGV3 A0A1L8E0P9 A0A182NLA0 A0A2M4AEF7 Q7QM15 A0A182IKX4 Q7Q9C1 A0A182HJJ2 A0A182UXX9 A0A2J7QDN1 A0A182M072 B4MRY0 Q28ZU9 B4LJC1 A0A1B0CZR5 B4JVZ0 A0A1A9URC8 A0A1A9Z2G6 B4P7M4 B3NN29 A0A3B0JNX9 T1PFB1 A0A1B0FK01 D1FQ06 A0A1W4WSQ1 A0A1L8EER1 A0A1A9WWU7 A0A1I8QAR8 A0A1A9YL53 A0A069DMX4 B3MCS3 B4NSC9 Q7JZK1 A0A1W4VCJ1 A0A0A9WBQ8 B4HN24 A0A0M4ETG2 N6U0D3 B4GGR1 B4KTT1 A0A0N7Z9G2 R4FPF4 U4U7N1 A0A161MLJ2 W8C2I8 A0A1B0C5E2 A0A0P4WG23 A0A2R7WG33 A0A0P5FVM7 A0A162QNW1 A0A0V0G6U0 A0A0L0C5I4 A0A0N8BP27 A0A0P6F0P7 A0A023F825 A0A0K8V405 A0A0P5F9X0 A0A0P5MI13 A0A0P5NJT7 A0A226EDV4 A0A034VMX7 D7EHX8 A0A3R7N0B5 A0A0P6JXI5 A0A0P5D5G1 A0A0P5KCS9 A0A0P5EQ33 A0A0P5M2C4 A0A0P5DPX8
PDB
5GUP
E-value=2.66468e-31,
Score=331
Ontologies
PATHWAY
PANTHER
Topology
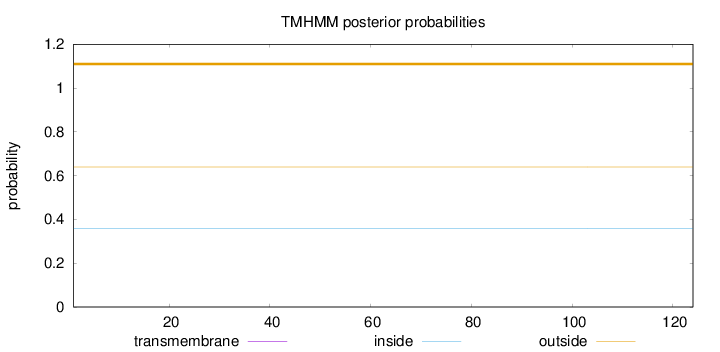
Length:
124
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.36018
outside
1 - 124
Population Genetic Test Statistics
Pi
319.901525
Theta
206.348953
Tajima's D
1.457888
CLR
0
CSRT
0.771661416929154
Interpretation
Uncertain
