Pre Gene Modal
BGIBMGA005460
Annotation
cell_division_protein_[Bombyx_mori]
Full name
Putative tRNA (cytidine(32)/guanosine(34)-2'-O)-methyltransferase
Alternative Name
2'-O-ribose RNA methyltransferase TRM7 homolog
Location in the cell
Cytoplasmic Reliability : 1.28 Extracellular Reliability : 1.482
Sequence
CDS
ATGGGCAAGACATCCAAAGATAAGCGAGATATATATTACAGGCTTGCTAAAGAAGAAGGTTGGCGTGCACGTAGCGCATTTAAACTTTTGCAAATTAACGAGGAATACAACATATTTAATGGTGTACTGCGTGCAGTGGATCTTTGCGCAGCACCAGGCAGTTGGAGTCAAGTTTTAACCAAGAATCTTCGACAAAATGCTGTAAATACTGAGGATGTTAAAATAGTTGCTGTTGATCTTCAGGCGATGGCAGCATTACCGGGAGTGAAACAAATTCAAGGGGATATTACCAAACAAGAAACAGCAAATGCTATAATAGAAGAGTTCCAAGGCCTCAAAGCAGATTTGGTTGTATGTGATGGTGCACCAGATGTAACAGGCTTACATGATATTGATGAATATGTCCAATCTCAGTTATTACTCGCTGCATTGAACATAACAACCCATGTTCTTAAGAATGGAGGTACATTTGTGGCCAAAATATTCAGAGGAAAAGATGTTTCCCTGCTGTACTCACAATTAAAACAGTTTTTTAAACTTGTGACAGTATCCAAACCTAGGAGTTCAAGAAATTCGAGCATTGAAGCCTTTGTAATTTGTGAGAAATACAATCCCCCAGAAGATTATGTGCCGAATATGGTCAATCCACTGTTAGATCATAAATACTTTGATTTGGATTCTGATTTTAATAGTTTTACTGGAATAAATAGATTTATAGTGCCATTCAATGCTTGTGGAGATCTTAGTGCCTATGACTCGGATACATCATATTCTCTACTGCTTGAAGGACAAACATCTTATGAGTATAAAAATCCAATACAAAGTCCTATCAATCCGCCTTACAAAGATGTTTTGGAAAAAACAAAAAAAATGAAAATAAATCATGTTCCTAAATAA
Protein
MGKTSKDKRDIYYRLAKEEGWRARSAFKLLQINEEYNIFNGVLRAVDLCAAPGSWSQVLTKNLRQNAVNTEDVKIVAVDLQAMAALPGVKQIQGDITKQETANAIIEEFQGLKADLVVCDGAPDVTGLHDIDEYVQSQLLLAALNITTHVLKNGGTFVAKIFRGKDVSLLYSQLKQFFKLVTVSKPRSSRNSSIEAFVICEKYNPPEDYVPNMVNPLLDHKYFDLDSDFNSFTGINRFIVPFNACGDLSAYDSDTSYSLLLEGQTSYEYKNPIQSPINPPYKDVLEKTKKMKINHVPK
Summary
Description
Methylates the 2'-O-ribose of nucleotides at positions 32 and 34 of the tRNA anticodon loop of substrate tRNAs.
Catalytic Activity
cytidine(32)/guanosine(34) in tRNA + 2 S-adenosyl-L-methionine = 2'-O-methylcytidine(32)/2'-O-methylguanosine(34) in tRNA + 2 H(+) + 2 S-adenosyl-L-homocysteine
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. RNA methyltransferase RlmE family. TRM7 subfamily.
Uniprot
H9J7G8
Q1HQ89
A0A2H1VBJ4
S4P3M9
A0A212EJG3
A0A2W1BXK8
+ More
A0A194QTY9 A0A194PN33 C9E269 A0A0L7LUQ1 D8VJ53 D8VJ30 D8VJ50 D8VJ10 D8VJ17 D8VJ21 D8VJ13 D8VJ23 D0VK13 D0VK11 D0VK20 D0VK33 D0VK27 D0VK21 D0VK25 D0VK24 A0A2P6KG06 T1J1K8 A0A0L8IES3 G3K9J1 G3K9M2 G3K9I3 G3K9L6 G3K9J7 G3K9K1 G3K9J2 A0A2L2YHR1 A0A2L2YFG8 G3K9K4 G3K9J4 G3K9L9 G3K9K7 G3K9J6 G3K9J9 G3K9I4 G3K9M0 G3K9L1 A0A1S3IAF9 A0A1S3H3X9 A0A087UZY3 A0A210R5C0 G3K9J3 K1QIU2 W5NFA4 G3K9L3 A0A3Q2FQ89 A0A3B4A3E6 V8P6T7 A0A2D4GYY0 A0A2D4HES9 A0A0K8RW81 A0A0F7Z2L4 A0A0B8RP91 A0A3B3CIC0 R7VKI1 G3K9J8 A0A1B6KG62 G3K9I8 G3K9L7 G3K9I9 A0A3B4A3W5 A0A3S1BMG4 A0A1B6CK10 A0A0P5KL34 A0A1B6F5K3 A0A0N8EGB1 C3ZIK7 A0A0P6G9D1 A0A0P5NZ93 A0A3B1JHN3 F6TQG1 A0A1Y1W4X9 A0A0N8CCT7 A0A0P6EZ21 A0A0B7AHG6 A0A3B4CFG4 A0A0B7AH47 E9G3S7 A0A3Q1GLJ2 Q4SE69 H3CYH4 V3Z4Z2 A0A2R7X0A0 W5UAG5 A0A2G5BBF3 A0A2G8JB06 A0A3B3H689 A0A1A8QZR8 A0A1A8L6L4 A0A1A8E5F1
A0A194QTY9 A0A194PN33 C9E269 A0A0L7LUQ1 D8VJ53 D8VJ30 D8VJ50 D8VJ10 D8VJ17 D8VJ21 D8VJ13 D8VJ23 D0VK13 D0VK11 D0VK20 D0VK33 D0VK27 D0VK21 D0VK25 D0VK24 A0A2P6KG06 T1J1K8 A0A0L8IES3 G3K9J1 G3K9M2 G3K9I3 G3K9L6 G3K9J7 G3K9K1 G3K9J2 A0A2L2YHR1 A0A2L2YFG8 G3K9K4 G3K9J4 G3K9L9 G3K9K7 G3K9J6 G3K9J9 G3K9I4 G3K9M0 G3K9L1 A0A1S3IAF9 A0A1S3H3X9 A0A087UZY3 A0A210R5C0 G3K9J3 K1QIU2 W5NFA4 G3K9L3 A0A3Q2FQ89 A0A3B4A3E6 V8P6T7 A0A2D4GYY0 A0A2D4HES9 A0A0K8RW81 A0A0F7Z2L4 A0A0B8RP91 A0A3B3CIC0 R7VKI1 G3K9J8 A0A1B6KG62 G3K9I8 G3K9L7 G3K9I9 A0A3B4A3W5 A0A3S1BMG4 A0A1B6CK10 A0A0P5KL34 A0A1B6F5K3 A0A0N8EGB1 C3ZIK7 A0A0P6G9D1 A0A0P5NZ93 A0A3B1JHN3 F6TQG1 A0A1Y1W4X9 A0A0N8CCT7 A0A0P6EZ21 A0A0B7AHG6 A0A3B4CFG4 A0A0B7AH47 E9G3S7 A0A3Q1GLJ2 Q4SE69 H3CYH4 V3Z4Z2 A0A2R7X0A0 W5UAG5 A0A2G5BBF3 A0A2G8JB06 A0A3B3H689 A0A1A8QZR8 A0A1A8L6L4 A0A1A8E5F1
EC Number
2.1.1.205
Pubmed
EMBL
BABH01013833
DQ443163
ABF51252.1
ODYU01001661
SOQ38195.1
GAIX01006374
+ More
JAA86186.1 AGBW02014484 OWR41598.1 KZ149904 PZC78385.1 KQ461154 KPJ08460.1 KQ459599 KPI94403.1 GQ452006 ACV83779.1 JTDY01000051 KOB79160.1 GQ987110 GQ987111 GQ987113 GQ987117 GQ987118 ADI81797.1 GQ987087 GQ987088 GQ987089 GQ987090 GQ987091 GQ987092 GQ987093 GQ987094 GQ987095 GQ987096 GQ987097 GQ987098 GQ987099 GQ987100 GQ987101 GQ987102 GQ987103 GQ987104 GQ987105 GQ987106 ADI81774.1 GQ987107 GQ987108 GQ987109 GQ987112 GQ987114 GQ987115 GQ987116 GQ987119 GQ987120 GQ987121 GQ987122 GQ987123 GQ987124 GQ987125 GQ987126 ADI81794.1 GQ987067 GQ987068 GQ987069 GQ987071 GQ987072 GQ987073 GQ987075 GQ987076 GQ987077 GQ987079 GQ987081 GQ987082 GQ987083 GQ987084 GQ987085 GQ987086 ADI81754.1 GQ987074 ADI81761.1 GQ987078 ADI81765.1 GQ987070 ADI81757.1 GQ987080 ADI81767.1 GU078084 GU078109 ACY09322.1 ACY09347.1 GU078082 GU078083 GU078085 GU078086 GU078087 GU078088 GU078089 GU078090 GU078105 GU078106 GU078107 GU078108 ACY09320.1 ACY09321.1 ACY09323.1 ACY09324.1 ACY09325.1 ACY09326.1 ACY09327.1 ACY09328.1 ACY09343.1 ACY09344.1 ACY09345.1 ACY09346.1 GU078091 GU078093 GU078094 GU078097 GU078099 GU078100 GU078101 GU078102 GU078103 ACY09329.1 ACY09331.1 ACY09332.1 ACY09335.1 ACY09337.1 ACY09338.1 ACY09339.1 ACY09340.1 ACY09341.1 GU078104 ACY09342.1 GU078098 ACY09336.1 GU078092 ACY09330.1 GU078096 ACY09334.1 GU078095 ACY09333.1 MWRG01012959 PRD25258.1 JH431789 KQ415942 KOF99535.1 JN174663 AEN68006.1 JN174694 AEN68037.1 JN174655 AEN67998.1 JN174677 JN174678 JN174680 JN174681 JN174682 JN174684 JN174687 JN174688 JN174693 AEN68020.1 AEN68021.1 AEN68023.1 AEN68024.1 AEN68025.1 AEN68027.1 AEN68030.1 AEN68031.1 AEN68036.1 JN174657 JN174658 JN174659 JN174662 JN174669 JN174672 JN174674 JN174675 AEN68000.1 AEN68001.1 AEN68002.1 AEN68005.1 AEN68012.1 AEN68015.1 AEN68017.1 AEN68018.1 JN174673 AEN68016.1 JN174664 AEN68007.1 IAAA01022333 LAA06765.1 IAAA01022334 LAA06767.1 JN174676 JN174686 AEN68019.1 AEN68029.1 JN174666 JN174667 AEN68009.1 AEN68010.1 JN174691 AEN68034.1 JN174679 AEN68022.1 JN174668 AEN68011.1 JN174671 AEN68014.1 JN174656 AEN67999.1 JN174690 JN174692 AEN68033.1 AEN68035.1 JN174683 AEN68026.1 KK122529 KFM82922.1 NEDP02000262 OWF56175.1 JN174665 AEN68008.1 JH823221 EKC36697.1 AHAT01031519 JN174685 AEN68028.1 AZIM01000635 ETE70035.1 IACJ01150332 LAA64924.1 IACK01015183 IACK01015185 LAA70463.1 GBKC01001375 JAG44695.1 GBEX01003709 JAI10851.1 GBSH01002397 JAG66629.1 AMQN01004037 KB292507 ELU17391.1 JN174670 AEN68013.1 GEBQ01029529 JAT10448.1 JN174660 AEN68003.1 JN174689 AEN68032.1 JN174661 AEN68004.1 RQTK01000007 RUS91682.1 GEDC01023464 JAS13834.1 GDIQ01208173 GDIQ01208172 GDIQ01168832 LRGB01001463 JAK43553.1 KZS11920.1 GECZ01024289 JAS45480.1 GDIQ01241558 GDIQ01240015 GDIQ01230263 GDIQ01209474 GDIQ01209473 GDIQ01161489 GDIQ01126149 GDIQ01101712 GDIQ01067823 GDIQ01029580 JAN65157.1 GG666627 EEN47732.1 GDIQ01039438 JAN55299.1 GDIQ01135800 JAL15926.1 EAAA01001403 MCFD01000009 ORX68581.1 GDIQ01092363 JAL59363.1 GDIQ01057275 JAN37462.1 HACG01032500 CEK79365.1 HACG01032501 CEK79366.1 GL732531 EFX85822.1 CAAE01014625 CAG01063.1 KB203251 ESO85773.1 KK856221 PTY25227.1 JT410164 AHH38953.1 KZ303500 PIA16322.1 MRZV01002950 PIK32928.1 HAEH01000349 HAEI01006702 SBR98708.1 HAEF01003073 HAEG01000064 SBR40455.1 HADZ01006064 HAEA01012718 SBQ41198.1
JAA86186.1 AGBW02014484 OWR41598.1 KZ149904 PZC78385.1 KQ461154 KPJ08460.1 KQ459599 KPI94403.1 GQ452006 ACV83779.1 JTDY01000051 KOB79160.1 GQ987110 GQ987111 GQ987113 GQ987117 GQ987118 ADI81797.1 GQ987087 GQ987088 GQ987089 GQ987090 GQ987091 GQ987092 GQ987093 GQ987094 GQ987095 GQ987096 GQ987097 GQ987098 GQ987099 GQ987100 GQ987101 GQ987102 GQ987103 GQ987104 GQ987105 GQ987106 ADI81774.1 GQ987107 GQ987108 GQ987109 GQ987112 GQ987114 GQ987115 GQ987116 GQ987119 GQ987120 GQ987121 GQ987122 GQ987123 GQ987124 GQ987125 GQ987126 ADI81794.1 GQ987067 GQ987068 GQ987069 GQ987071 GQ987072 GQ987073 GQ987075 GQ987076 GQ987077 GQ987079 GQ987081 GQ987082 GQ987083 GQ987084 GQ987085 GQ987086 ADI81754.1 GQ987074 ADI81761.1 GQ987078 ADI81765.1 GQ987070 ADI81757.1 GQ987080 ADI81767.1 GU078084 GU078109 ACY09322.1 ACY09347.1 GU078082 GU078083 GU078085 GU078086 GU078087 GU078088 GU078089 GU078090 GU078105 GU078106 GU078107 GU078108 ACY09320.1 ACY09321.1 ACY09323.1 ACY09324.1 ACY09325.1 ACY09326.1 ACY09327.1 ACY09328.1 ACY09343.1 ACY09344.1 ACY09345.1 ACY09346.1 GU078091 GU078093 GU078094 GU078097 GU078099 GU078100 GU078101 GU078102 GU078103 ACY09329.1 ACY09331.1 ACY09332.1 ACY09335.1 ACY09337.1 ACY09338.1 ACY09339.1 ACY09340.1 ACY09341.1 GU078104 ACY09342.1 GU078098 ACY09336.1 GU078092 ACY09330.1 GU078096 ACY09334.1 GU078095 ACY09333.1 MWRG01012959 PRD25258.1 JH431789 KQ415942 KOF99535.1 JN174663 AEN68006.1 JN174694 AEN68037.1 JN174655 AEN67998.1 JN174677 JN174678 JN174680 JN174681 JN174682 JN174684 JN174687 JN174688 JN174693 AEN68020.1 AEN68021.1 AEN68023.1 AEN68024.1 AEN68025.1 AEN68027.1 AEN68030.1 AEN68031.1 AEN68036.1 JN174657 JN174658 JN174659 JN174662 JN174669 JN174672 JN174674 JN174675 AEN68000.1 AEN68001.1 AEN68002.1 AEN68005.1 AEN68012.1 AEN68015.1 AEN68017.1 AEN68018.1 JN174673 AEN68016.1 JN174664 AEN68007.1 IAAA01022333 LAA06765.1 IAAA01022334 LAA06767.1 JN174676 JN174686 AEN68019.1 AEN68029.1 JN174666 JN174667 AEN68009.1 AEN68010.1 JN174691 AEN68034.1 JN174679 AEN68022.1 JN174668 AEN68011.1 JN174671 AEN68014.1 JN174656 AEN67999.1 JN174690 JN174692 AEN68033.1 AEN68035.1 JN174683 AEN68026.1 KK122529 KFM82922.1 NEDP02000262 OWF56175.1 JN174665 AEN68008.1 JH823221 EKC36697.1 AHAT01031519 JN174685 AEN68028.1 AZIM01000635 ETE70035.1 IACJ01150332 LAA64924.1 IACK01015183 IACK01015185 LAA70463.1 GBKC01001375 JAG44695.1 GBEX01003709 JAI10851.1 GBSH01002397 JAG66629.1 AMQN01004037 KB292507 ELU17391.1 JN174670 AEN68013.1 GEBQ01029529 JAT10448.1 JN174660 AEN68003.1 JN174689 AEN68032.1 JN174661 AEN68004.1 RQTK01000007 RUS91682.1 GEDC01023464 JAS13834.1 GDIQ01208173 GDIQ01208172 GDIQ01168832 LRGB01001463 JAK43553.1 KZS11920.1 GECZ01024289 JAS45480.1 GDIQ01241558 GDIQ01240015 GDIQ01230263 GDIQ01209474 GDIQ01209473 GDIQ01161489 GDIQ01126149 GDIQ01101712 GDIQ01067823 GDIQ01029580 JAN65157.1 GG666627 EEN47732.1 GDIQ01039438 JAN55299.1 GDIQ01135800 JAL15926.1 EAAA01001403 MCFD01000009 ORX68581.1 GDIQ01092363 JAL59363.1 GDIQ01057275 JAN37462.1 HACG01032500 CEK79365.1 HACG01032501 CEK79366.1 GL732531 EFX85822.1 CAAE01014625 CAG01063.1 KB203251 ESO85773.1 KK856221 PTY25227.1 JT410164 AHH38953.1 KZ303500 PIA16322.1 MRZV01002950 PIK32928.1 HAEH01000349 HAEI01006702 SBR98708.1 HAEF01003073 HAEG01000064 SBR40455.1 HADZ01006064 HAEA01012718 SBQ41198.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000037510
UP000053454
+ More
UP000085678 UP000054359 UP000242188 UP000005408 UP000018468 UP000265020 UP000261520 UP000261560 UP000014760 UP000271974 UP000076858 UP000001554 UP000018467 UP000008144 UP000193922 UP000261440 UP000000305 UP000257200 UP000007303 UP000030746 UP000221080 UP000242474 UP000230750 UP000001038
UP000085678 UP000054359 UP000242188 UP000005408 UP000018468 UP000265020 UP000261520 UP000261560 UP000014760 UP000271974 UP000076858 UP000001554 UP000018467 UP000008144 UP000193922 UP000261440 UP000000305 UP000257200 UP000007303 UP000030746 UP000221080 UP000242474 UP000230750 UP000001038
Pfam
PF01728 FtsJ
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9J7G8
Q1HQ89
A0A2H1VBJ4
S4P3M9
A0A212EJG3
A0A2W1BXK8
+ More
A0A194QTY9 A0A194PN33 C9E269 A0A0L7LUQ1 D8VJ53 D8VJ30 D8VJ50 D8VJ10 D8VJ17 D8VJ21 D8VJ13 D8VJ23 D0VK13 D0VK11 D0VK20 D0VK33 D0VK27 D0VK21 D0VK25 D0VK24 A0A2P6KG06 T1J1K8 A0A0L8IES3 G3K9J1 G3K9M2 G3K9I3 G3K9L6 G3K9J7 G3K9K1 G3K9J2 A0A2L2YHR1 A0A2L2YFG8 G3K9K4 G3K9J4 G3K9L9 G3K9K7 G3K9J6 G3K9J9 G3K9I4 G3K9M0 G3K9L1 A0A1S3IAF9 A0A1S3H3X9 A0A087UZY3 A0A210R5C0 G3K9J3 K1QIU2 W5NFA4 G3K9L3 A0A3Q2FQ89 A0A3B4A3E6 V8P6T7 A0A2D4GYY0 A0A2D4HES9 A0A0K8RW81 A0A0F7Z2L4 A0A0B8RP91 A0A3B3CIC0 R7VKI1 G3K9J8 A0A1B6KG62 G3K9I8 G3K9L7 G3K9I9 A0A3B4A3W5 A0A3S1BMG4 A0A1B6CK10 A0A0P5KL34 A0A1B6F5K3 A0A0N8EGB1 C3ZIK7 A0A0P6G9D1 A0A0P5NZ93 A0A3B1JHN3 F6TQG1 A0A1Y1W4X9 A0A0N8CCT7 A0A0P6EZ21 A0A0B7AHG6 A0A3B4CFG4 A0A0B7AH47 E9G3S7 A0A3Q1GLJ2 Q4SE69 H3CYH4 V3Z4Z2 A0A2R7X0A0 W5UAG5 A0A2G5BBF3 A0A2G8JB06 A0A3B3H689 A0A1A8QZR8 A0A1A8L6L4 A0A1A8E5F1
A0A194QTY9 A0A194PN33 C9E269 A0A0L7LUQ1 D8VJ53 D8VJ30 D8VJ50 D8VJ10 D8VJ17 D8VJ21 D8VJ13 D8VJ23 D0VK13 D0VK11 D0VK20 D0VK33 D0VK27 D0VK21 D0VK25 D0VK24 A0A2P6KG06 T1J1K8 A0A0L8IES3 G3K9J1 G3K9M2 G3K9I3 G3K9L6 G3K9J7 G3K9K1 G3K9J2 A0A2L2YHR1 A0A2L2YFG8 G3K9K4 G3K9J4 G3K9L9 G3K9K7 G3K9J6 G3K9J9 G3K9I4 G3K9M0 G3K9L1 A0A1S3IAF9 A0A1S3H3X9 A0A087UZY3 A0A210R5C0 G3K9J3 K1QIU2 W5NFA4 G3K9L3 A0A3Q2FQ89 A0A3B4A3E6 V8P6T7 A0A2D4GYY0 A0A2D4HES9 A0A0K8RW81 A0A0F7Z2L4 A0A0B8RP91 A0A3B3CIC0 R7VKI1 G3K9J8 A0A1B6KG62 G3K9I8 G3K9L7 G3K9I9 A0A3B4A3W5 A0A3S1BMG4 A0A1B6CK10 A0A0P5KL34 A0A1B6F5K3 A0A0N8EGB1 C3ZIK7 A0A0P6G9D1 A0A0P5NZ93 A0A3B1JHN3 F6TQG1 A0A1Y1W4X9 A0A0N8CCT7 A0A0P6EZ21 A0A0B7AHG6 A0A3B4CFG4 A0A0B7AH47 E9G3S7 A0A3Q1GLJ2 Q4SE69 H3CYH4 V3Z4Z2 A0A2R7X0A0 W5UAG5 A0A2G5BBF3 A0A2G8JB06 A0A3B3H689 A0A1A8QZR8 A0A1A8L6L4 A0A1A8E5F1
PDB
6EM5
E-value=6.21612e-38,
Score=394
Ontologies
GO
PANTHER
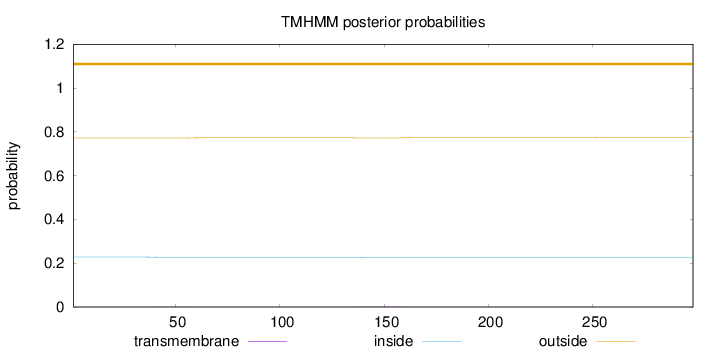
Topology
Subcellular location
Cytoplasm
Length:
298
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06431
Exp number, first 60 AAs:
0.02444
Total prob of N-in:
0.22862
outside
1 - 298
Population Genetic Test Statistics
Pi
280.541301
Theta
196.995234
Tajima's D
1.240209
CLR
0.722697
CSRT
0.72306384680766
Interpretation
Uncertain
