Gene
KWMTBOMO04530
Pre Gene Modal
BGIBMGA005459
Annotation
PREDICTED:_putative_methyltransferase_NSUN6_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.352
Sequence
CDS
ATGGAGACGCCTAAATATACAACATTTCGTGTAAACATATTAAAAAAAAATCACCATGAAAGTTTACAACAATACCTCTTGCAACAAGCCGATGTATTTGCTACCAAGCAGATCCCTAGCTTTTACTTTTTGAAACCTGACTGTCTTATATTAGAAAGATGGTCTGAAAGTGTAATAGTGGATCCAACTGGGAAGGAGGTGATTGTTGATGCATTATGTGCTGCTGCAGTATTAAGGGGTGCTCATGTGTTTGCTCCAGGAGTAATGGGTCTTCCAAGCAGTCTCCATTTAGATGAAAAAGTAGACATTTATGGTGACCTTGAAAAGAAATGCAAAAGAGGATTGAAATCGTTGTATGATGGGAAGAAGAAATATGTGGGTACGGGATATTTAAAAATGTTGCGGTCCGAATTATTTGATAATGGAATCAAGCCAAGTGGTATCGCTGTGCATACTTTAATGCCTGTGTCTAGATTGCCAGTTGTGAATGAATCTGTCTGTCCTAAAGGTGAACTTCTCTTACAAAACTTACCATCTATTGTATGCGGTTGGGTAGTCAATGCACAATCTAATGAATACATACTCGACATGTGTGCTGCTCCAGGCAACAAAACAACACATCTGGCTGAGATGTCTAAAAATCAGGCATGCATCACAGCATTAGATAAAACATTAGAAAAAACAGAAAGGATTCGAGAAACCTGTCAAATCCAAGGTGTTACTTGCGTCACAGCTTATGCATTTGATGCCACCAAGTGTCATTCGGAAGAAAGTCTTGGAATAGCAAATGGACCCCCATTTCCATCGAATTGTTTCGATAAAATACTCCTTGATGCGCCTTGTAGTGGCTTGGGGCAACGACCTCAATTAATAAACATGATGTCACCGAAAATGATAAAATCCTACAAATTCCTACAACGTAAACTGATAAGTGAGGCTGTAAAACTATTGAAAGTTGGTGGTAAATTAGTTTACAGTACCTGTACTATAACTGAAGATGAAAATGAAGGTCTGGTGTCATGGGTGCTAGAAAAGTTTCCATGTTTAAAACTTATACCTGCTGAACCACTCCTCGGAGGGCCTGGTCTACCTAATTTAGGTCTTGATGATGAACAGAGAAAAATGGTGCAACGCTTTGGGCCTCACATAGATGCGTTACGACCTGCAGATGAAATATATCGAGATAGTATAGGGTTTTTCATAGCAGCCTTTACGAAAACGGCAAATAATAAATAA
Protein
METPKYTTFRVNILKKNHHESLQQYLLQQADVFATKQIPSFYFLKPDCLILERWSESVIVDPTGKEVIVDALCAAAVLRGAHVFAPGVMGLPSSLHLDEKVDIYGDLEKKCKRGLKSLYDGKKKYVGTGYLKMLRSELFDNGIKPSGIAVHTLMPVSRLPVVNESVCPKGELLLQNLPSIVCGWVVNAQSNEYILDMCAAPGNKTTHLAEMSKNQACITALDKTLEKTERIRETCQIQGVTCVTAYAFDATKCHSEESLGIANGPPFPSNCFDKILLDAPCSGLGQRPQLINMMSPKMIKSYKFLQRKLISEAVKLLKVGGKLVYSTCTITEDENEGLVSWVLEKFPCLKLIPAEPLLGGPGLPNLGLDDEQRKMVQRFGPHIDALRPADEIYRDSIGFFIAAFTKTANNK
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. RsmB/NOP family.
Uniprot
H9J7G7
A0A2H1VFG8
A0A3S2P3M9
A0A2W1BXL6
A0A212EJD2
A0A194PTE5
+ More
A0A067RP03 A0A2P8XXH3 A0A069DTH6 A0A023F122 K7IVF7 A0A1I8NU66 N6UMA3 A0A232EX71 A0A1I8NU67 Q2M0P6 A0A0P4VL17 B4GRB9 A0A1I8NU64 A0A224XPU0 A0A1I8MCJ3 A0A1I8MCJ2 N6UR01 A0A0L0CRX7 A0A1W4UR86 B4J3R9 A0A3M6T4N1 A0A0Q9XCV4 E2BY39 A0A0M3QVU9 C3ZX25 B4KYK9 A0A034WKL7 F4X4X2 A0A151I7G5 A0A0J7KDX9 A0A034WJQ9 A0A0K8WFA8 B4QLB6 A0A026X3C3 D6WZC9 A7SEM2 E2AW30 A0A0K8VHG3 A0A158NVW8 A0A195ETB1 B3MB98 A0A151HXW0 A0A0K8UDB5 A0A2B4RRT0 A0A1A9Z3M2 R7VCP3 B4IB54 T1JE95 A0A1A9UXV3 A0A3B0JZ89 A0A3Q1HN61 Q8IPS4 E6ZJ27 A0A315VGD8 B4LDP6 B3NEG7 A0A3B3XWD0 A0A0L8HSL2 A0A3Q3K8H2 A0A2I4BM21 A0A3Q2UGV7 A0A3B3UVF9 W5MY52 A0A087YFC1 A0A146NTV8 A0A146YC04 A0A1A9WS75 E9IB25 V5HYV2 A0A0R4IAT4 B4PIR2 A0A1Y1K8H0 A0A3B4TVQ1 F7BYX0 V5HTK3 A0A3B4YHC1 A0A3B5LVQ1 A0A3Q1GHJ6 A0A2R5L697 A0A3P8N8S4 W4XEF7 A0A3P9AUV7 A0A091NAL7 A0A3Q1CQD0 A0A3B3BAV9 A0A3Q2VHZ7 A0A2H8TM09 A0A3P9M2X7 H2M5A7 A0A3B3I1V2 A0A3P9NRD9 A0A1A8CGU9
A0A067RP03 A0A2P8XXH3 A0A069DTH6 A0A023F122 K7IVF7 A0A1I8NU66 N6UMA3 A0A232EX71 A0A1I8NU67 Q2M0P6 A0A0P4VL17 B4GRB9 A0A1I8NU64 A0A224XPU0 A0A1I8MCJ3 A0A1I8MCJ2 N6UR01 A0A0L0CRX7 A0A1W4UR86 B4J3R9 A0A3M6T4N1 A0A0Q9XCV4 E2BY39 A0A0M3QVU9 C3ZX25 B4KYK9 A0A034WKL7 F4X4X2 A0A151I7G5 A0A0J7KDX9 A0A034WJQ9 A0A0K8WFA8 B4QLB6 A0A026X3C3 D6WZC9 A7SEM2 E2AW30 A0A0K8VHG3 A0A158NVW8 A0A195ETB1 B3MB98 A0A151HXW0 A0A0K8UDB5 A0A2B4RRT0 A0A1A9Z3M2 R7VCP3 B4IB54 T1JE95 A0A1A9UXV3 A0A3B0JZ89 A0A3Q1HN61 Q8IPS4 E6ZJ27 A0A315VGD8 B4LDP6 B3NEG7 A0A3B3XWD0 A0A0L8HSL2 A0A3Q3K8H2 A0A2I4BM21 A0A3Q2UGV7 A0A3B3UVF9 W5MY52 A0A087YFC1 A0A146NTV8 A0A146YC04 A0A1A9WS75 E9IB25 V5HYV2 A0A0R4IAT4 B4PIR2 A0A1Y1K8H0 A0A3B4TVQ1 F7BYX0 V5HTK3 A0A3B4YHC1 A0A3B5LVQ1 A0A3Q1GHJ6 A0A2R5L697 A0A3P8N8S4 W4XEF7 A0A3P9AUV7 A0A091NAL7 A0A3Q1CQD0 A0A3B3BAV9 A0A3Q2VHZ7 A0A2H8TM09 A0A3P9M2X7 H2M5A7 A0A3B3I1V2 A0A3P9NRD9 A0A1A8CGU9
Pubmed
19121390
28756777
22118469
26354079
24845553
29403074
+ More
26334808 25474469 20075255 23537049 28648823 15632085 17994087 27129103 25315136 26108605 30382153 20798317 18563158 25348373 21719571 22936249 24508170 30249741 18362917 19820115 17615350 21347285 23254933 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29703783 21282665 25765539 23594743 17550304 28004739 18464734 25186727 29451363 17554307
26334808 25474469 20075255 23537049 28648823 15632085 17994087 27129103 25315136 26108605 30382153 20798317 18563158 25348373 21719571 22936249 24508170 30249741 18362917 19820115 17615350 21347285 23254933 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29703783 21282665 25765539 23594743 17550304 28004739 18464734 25186727 29451363 17554307
EMBL
BABH01013829
ODYU01002287
SOQ39575.1
RSAL01000032
RVE51548.1
KZ149904
+ More
PZC78384.1 AGBW02014484 OWR41596.1 KQ459599 KPI94405.1 KK852510 KDR22345.1 PYGN01001200 PSN36710.1 GBGD01001644 JAC87245.1 GBBI01003734 JAC14978.1 APGK01009148 KB737869 KB632374 ENN82870.1 ERL93799.1 NNAY01001775 OXU22955.1 CH379069 EAL30884.2 GDKW01001511 JAI55084.1 CH479188 EDW40304.1 GFTR01005844 JAW10582.1 APGK01005562 KB736320 ENN83221.1 JRES01000005 KNC34976.1 CH916366 EDV97300.1 RCHS01004337 RMX35937.1 CH933809 KRG06379.1 GL451420 EFN79318.1 CP012525 ALC43033.1 GG666707 EEN42903.1 EDW18820.1 GAKP01004045 JAC54907.1 GL888679 EGI58545.1 KQ978426 KYM94030.1 LBMM01008741 KMQ88658.1 GAKP01004043 JAC54909.1 GDHF01002547 JAI49767.1 CM000363 CM002912 EDX11534.1 KMZ01248.1 KK107020 QOIP01000006 EZA62583.1 RLU21849.1 KQ971372 EFA09726.1 DS469638 EDO37813.1 GL443213 EFN62452.1 GDHF01014016 GDHF01002427 JAI38298.1 JAI49887.1 ADTU01027564 KQ981993 KYN31119.1 CH902618 EDV41399.2 KQ976730 KYM76407.1 GDHF01027992 GDHF01027685 GDHF01022661 JAI24322.1 JAI24629.1 JAI29653.1 LSMT01000380 PFX19038.1 AMQN01000610 KB293180 ELU16404.1 CH480826 EDW44517.1 JH432116 OUUW01000012 SPP87395.1 AE014296 BT046128 AAN12208.1 ACI46516.1 FQ310508 CBN82061.1 NHOQ01002355 PWA18182.1 CH940647 EDW70007.2 CH954178 EDV52802.1 KQ417370 KOF92253.1 AHAT01017623 AYCK01006251 GCES01151129 JAQ35193.1 GCES01034644 JAR51679.1 GL762106 EFZ22220.1 GANP01002746 JAB81722.1 CU607104 CM000159 EDW95570.2 GEZM01091879 JAV56728.1 AAPN01114285 AAPN01114286 GANP01002744 JAB81724.1 GGLE01000900 MBY05026.1 AAGJ04029351 KK844920 KFP86728.1 GFXV01003176 MBW14981.1 HADZ01014065 SBP78006.1
PZC78384.1 AGBW02014484 OWR41596.1 KQ459599 KPI94405.1 KK852510 KDR22345.1 PYGN01001200 PSN36710.1 GBGD01001644 JAC87245.1 GBBI01003734 JAC14978.1 APGK01009148 KB737869 KB632374 ENN82870.1 ERL93799.1 NNAY01001775 OXU22955.1 CH379069 EAL30884.2 GDKW01001511 JAI55084.1 CH479188 EDW40304.1 GFTR01005844 JAW10582.1 APGK01005562 KB736320 ENN83221.1 JRES01000005 KNC34976.1 CH916366 EDV97300.1 RCHS01004337 RMX35937.1 CH933809 KRG06379.1 GL451420 EFN79318.1 CP012525 ALC43033.1 GG666707 EEN42903.1 EDW18820.1 GAKP01004045 JAC54907.1 GL888679 EGI58545.1 KQ978426 KYM94030.1 LBMM01008741 KMQ88658.1 GAKP01004043 JAC54909.1 GDHF01002547 JAI49767.1 CM000363 CM002912 EDX11534.1 KMZ01248.1 KK107020 QOIP01000006 EZA62583.1 RLU21849.1 KQ971372 EFA09726.1 DS469638 EDO37813.1 GL443213 EFN62452.1 GDHF01014016 GDHF01002427 JAI38298.1 JAI49887.1 ADTU01027564 KQ981993 KYN31119.1 CH902618 EDV41399.2 KQ976730 KYM76407.1 GDHF01027992 GDHF01027685 GDHF01022661 JAI24322.1 JAI24629.1 JAI29653.1 LSMT01000380 PFX19038.1 AMQN01000610 KB293180 ELU16404.1 CH480826 EDW44517.1 JH432116 OUUW01000012 SPP87395.1 AE014296 BT046128 AAN12208.1 ACI46516.1 FQ310508 CBN82061.1 NHOQ01002355 PWA18182.1 CH940647 EDW70007.2 CH954178 EDV52802.1 KQ417370 KOF92253.1 AHAT01017623 AYCK01006251 GCES01151129 JAQ35193.1 GCES01034644 JAR51679.1 GL762106 EFZ22220.1 GANP01002746 JAB81722.1 CU607104 CM000159 EDW95570.2 GEZM01091879 JAV56728.1 AAPN01114285 AAPN01114286 GANP01002744 JAB81724.1 GGLE01000900 MBY05026.1 AAGJ04029351 KK844920 KFP86728.1 GFXV01003176 MBW14981.1 HADZ01014065 SBP78006.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000027135
UP000245037
+ More
UP000002358 UP000095300 UP000019118 UP000030742 UP000215335 UP000001819 UP000008744 UP000095301 UP000037069 UP000192221 UP000001070 UP000275408 UP000009192 UP000008237 UP000092553 UP000001554 UP000007755 UP000078542 UP000036403 UP000000304 UP000053097 UP000279307 UP000007266 UP000001593 UP000000311 UP000005205 UP000078541 UP000007801 UP000078540 UP000225706 UP000092445 UP000014760 UP000001292 UP000078200 UP000268350 UP000265040 UP000000803 UP000008792 UP000008711 UP000261480 UP000053454 UP000261600 UP000192220 UP000265000 UP000261500 UP000018468 UP000028760 UP000091820 UP000000437 UP000002282 UP000261420 UP000002279 UP000261360 UP000261380 UP000257200 UP000265100 UP000007110 UP000265160 UP000257160 UP000261560 UP000264840 UP000265180 UP000001038 UP000242638
UP000002358 UP000095300 UP000019118 UP000030742 UP000215335 UP000001819 UP000008744 UP000095301 UP000037069 UP000192221 UP000001070 UP000275408 UP000009192 UP000008237 UP000092553 UP000001554 UP000007755 UP000078542 UP000036403 UP000000304 UP000053097 UP000279307 UP000007266 UP000001593 UP000000311 UP000005205 UP000078541 UP000007801 UP000078540 UP000225706 UP000092445 UP000014760 UP000001292 UP000078200 UP000268350 UP000265040 UP000000803 UP000008792 UP000008711 UP000261480 UP000053454 UP000261600 UP000192220 UP000265000 UP000261500 UP000018468 UP000028760 UP000091820 UP000000437 UP000002282 UP000261420 UP000002279 UP000261360 UP000261380 UP000257200 UP000265100 UP000007110 UP000265160 UP000257160 UP000261560 UP000264840 UP000265180 UP000001038 UP000242638
Interpro
Gene 3D
ProteinModelPortal
H9J7G7
A0A2H1VFG8
A0A3S2P3M9
A0A2W1BXL6
A0A212EJD2
A0A194PTE5
+ More
A0A067RP03 A0A2P8XXH3 A0A069DTH6 A0A023F122 K7IVF7 A0A1I8NU66 N6UMA3 A0A232EX71 A0A1I8NU67 Q2M0P6 A0A0P4VL17 B4GRB9 A0A1I8NU64 A0A224XPU0 A0A1I8MCJ3 A0A1I8MCJ2 N6UR01 A0A0L0CRX7 A0A1W4UR86 B4J3R9 A0A3M6T4N1 A0A0Q9XCV4 E2BY39 A0A0M3QVU9 C3ZX25 B4KYK9 A0A034WKL7 F4X4X2 A0A151I7G5 A0A0J7KDX9 A0A034WJQ9 A0A0K8WFA8 B4QLB6 A0A026X3C3 D6WZC9 A7SEM2 E2AW30 A0A0K8VHG3 A0A158NVW8 A0A195ETB1 B3MB98 A0A151HXW0 A0A0K8UDB5 A0A2B4RRT0 A0A1A9Z3M2 R7VCP3 B4IB54 T1JE95 A0A1A9UXV3 A0A3B0JZ89 A0A3Q1HN61 Q8IPS4 E6ZJ27 A0A315VGD8 B4LDP6 B3NEG7 A0A3B3XWD0 A0A0L8HSL2 A0A3Q3K8H2 A0A2I4BM21 A0A3Q2UGV7 A0A3B3UVF9 W5MY52 A0A087YFC1 A0A146NTV8 A0A146YC04 A0A1A9WS75 E9IB25 V5HYV2 A0A0R4IAT4 B4PIR2 A0A1Y1K8H0 A0A3B4TVQ1 F7BYX0 V5HTK3 A0A3B4YHC1 A0A3B5LVQ1 A0A3Q1GHJ6 A0A2R5L697 A0A3P8N8S4 W4XEF7 A0A3P9AUV7 A0A091NAL7 A0A3Q1CQD0 A0A3B3BAV9 A0A3Q2VHZ7 A0A2H8TM09 A0A3P9M2X7 H2M5A7 A0A3B3I1V2 A0A3P9NRD9 A0A1A8CGU9
A0A067RP03 A0A2P8XXH3 A0A069DTH6 A0A023F122 K7IVF7 A0A1I8NU66 N6UMA3 A0A232EX71 A0A1I8NU67 Q2M0P6 A0A0P4VL17 B4GRB9 A0A1I8NU64 A0A224XPU0 A0A1I8MCJ3 A0A1I8MCJ2 N6UR01 A0A0L0CRX7 A0A1W4UR86 B4J3R9 A0A3M6T4N1 A0A0Q9XCV4 E2BY39 A0A0M3QVU9 C3ZX25 B4KYK9 A0A034WKL7 F4X4X2 A0A151I7G5 A0A0J7KDX9 A0A034WJQ9 A0A0K8WFA8 B4QLB6 A0A026X3C3 D6WZC9 A7SEM2 E2AW30 A0A0K8VHG3 A0A158NVW8 A0A195ETB1 B3MB98 A0A151HXW0 A0A0K8UDB5 A0A2B4RRT0 A0A1A9Z3M2 R7VCP3 B4IB54 T1JE95 A0A1A9UXV3 A0A3B0JZ89 A0A3Q1HN61 Q8IPS4 E6ZJ27 A0A315VGD8 B4LDP6 B3NEG7 A0A3B3XWD0 A0A0L8HSL2 A0A3Q3K8H2 A0A2I4BM21 A0A3Q2UGV7 A0A3B3UVF9 W5MY52 A0A087YFC1 A0A146NTV8 A0A146YC04 A0A1A9WS75 E9IB25 V5HYV2 A0A0R4IAT4 B4PIR2 A0A1Y1K8H0 A0A3B4TVQ1 F7BYX0 V5HTK3 A0A3B4YHC1 A0A3B5LVQ1 A0A3Q1GHJ6 A0A2R5L697 A0A3P8N8S4 W4XEF7 A0A3P9AUV7 A0A091NAL7 A0A3Q1CQD0 A0A3B3BAV9 A0A3Q2VHZ7 A0A2H8TM09 A0A3P9M2X7 H2M5A7 A0A3B3I1V2 A0A3P9NRD9 A0A1A8CGU9
PDB
5WWT
E-value=5.53672e-69,
Score=663
Ontologies
KEGG
GO
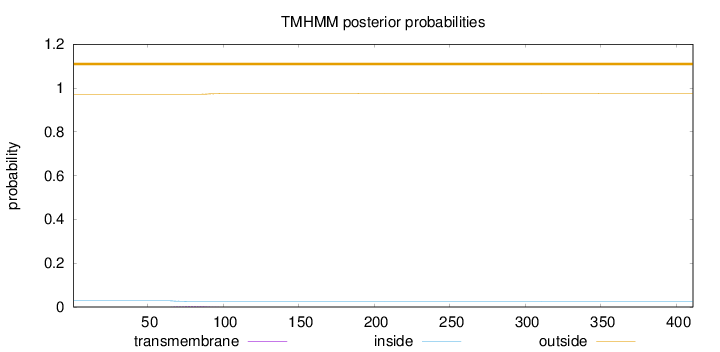
Topology
Length:
411
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0701000000000001
Exp number, first 60 AAs:
0.00043
Total prob of N-in:
0.02832
outside
1 - 411
Population Genetic Test Statistics
Pi
231.307634
Theta
187.162748
Tajima's D
0.98615
CLR
26.465379
CSRT
0.657567121643918
Interpretation
Uncertain
