Pre Gene Modal
BGIBMGA005458
Annotation
PREDICTED:_serine--pyruvate_aminotransferase-like_[Amyelois_transitella]
Full name
Serine--pyruvate aminotransferase
Alternative Name
Alanine--glyoxylate aminotransferase
Location in the cell
Extracellular Reliability : 0.876
Sequence
CDS
ATGACCGAAATTAAACTAACTGTTCAAGCTCCCACATTAGTAAATCGAGACTTTGTCAAGCCCACGCTGTGTGGTCCAGGCCCCTGTGACGTCTGGCCCTCAGTTAATGAGGCTCTCACCAAACCAGTTATTTCGCCAATTTGCGATGAACATTTCTACGTACTAGATGACATTCGCGCCGGGTTGAAGTATGTTTTTCAGACAAACTGCGACACAGTTCTCGCCCTCAGTGGATCTGGTCACGGTGGTATGGAGACTGTTATAAATAATTTGGTCGGACCCGGTGAAACTTTGCTGGTAGCCAAGAGGGGCTTTTGGTGTGAGCGTGCTCTTGAAATGGCTAAGAGATACGGAATCAACACTATAACAACGACTGTACCGATGAACGAGACGTTTAGTTTGCAACAGCTAGAAAATGAGCTAAGAAAACACCGTCCAACCGCTTTGTTTATAGTCCACGGCGATTCATCTACTGGAATAGTACAAAACTTGAAAAGTGTGGGCGAATTATGTCAAAGATATGGCACCCTTTTGCTGGTCGATACGGTAGTGTCGCTGGTTGGTGTACCATTCTTTATGGACGATTGGAAAATAGACGCGGTTTATACTTCAACCCAAAAAGCTTTCAGCGGCCCAGCTGGCATCAGTCCTGTAGCTTTCAGTGATCGTGCTCTGAACAAAATCAACAACAGAACTCATCAACCTCCTTTCTATTTTGACGTCAAACTATTGAGCCAACAATGGAATTGTTACGGTAGTACAAGAGTATATCATCACACTCTAAGTTCTCCGCTATTGTGGGCGCTACGGCAATGTCTGCAGGAAGTCATCAAAGAGACTTTACCCAGATCTTGGGCCCGACATGCTGCAACCACTAAACACTTCCTCAAAAGAGTAGAACAGCTCTCCTTAAAGACCTTTGTACCGAAACCGGAGGATCGCTTAATGACCGTCACAACGGTAGTCTTGCCCAAAGGATATGATTATTTACAATTCTCTAAATATATGAGAGAGAAATACAATATCTTAATATTTCCCGGCATCGGTCAAACTGTAGGTAAAACCCTACGTATTGGACTTATGGGTGTCAACTCTACAATACAAGTCGCCGATGCTGTTGCAGACGCTGTGGCAGATTGA
Protein
MTEIKLTVQAPTLVNRDFVKPTLCGPGPCDVWPSVNEALTKPVISPICDEHFYVLDDIRAGLKYVFQTNCDTVLALSGSGHGGMETVINNLVGPGETLLVAKRGFWCERALEMAKRYGINTITTTVPMNETFSLQQLENELRKHRPTALFIVHGDSSTGIVQNLKSVGELCQRYGTLLLVDTVVSLVGVPFFMDDWKIDAVYTSTQKAFSGPAGISPVAFSDRALNKINNRTHQPPFYFDVKLLSQQWNCYGSTRVYHHTLSSPLLWALRQCLQEVIKETLPRSWARHAATTKHFLKRVEQLSLKTFVPKPEDRLMTVTTVVLPKGYDYLQFSKYMREKYNILIFPGIGQTVGKTLRIGLMGVNSTIQVADAVADAVAD
Summary
Catalytic Activity
L-serine + pyruvate = 3-hydroxypyruvate + L-alanine
glyoxylate + L-alanine = glycine + pyruvate
glyoxylate + L-alanine = glycine + pyruvate
Cofactor
pyridoxal 5'-phosphate
Similarity
Belongs to the class-V pyridoxal-phosphate-dependent aminotransferase family.
Feature
chain Serine--pyruvate aminotransferase
Uniprot
H9J7G6
A0A2A4JA13
A0A2W1BTN4
A0A2W1BTI9
A0A2A4J9J0
A0A212F566
+ More
A0A3S2N685 A0A2H1VZD4 A0A194PTF0 A0A2H1W814 A0A194QT08 A0A194PNT9 A0A0L7LVG8 D6WZD5 A0A1D2M7D5 A0A2J7QYG5 B4MTD7 U5EHZ8 A0A0M9ABU5 A0A0P4VW80 A0A0P4W285 A0A182NL11 V5IAQ9 B3MZ47 A0A1W4VFD6 Q9W3Z3 C6TP18 B3NXS8 A0A3B0KL32 T1PAB8 B4PYT0 B4R5M1 A0A3Q4MTI7 A0A3Q2WPW6 Q29J54 B4GTB5 A0A3B4GVH1 A0A087SQK2 A0A3P8RB99 A0A3P9BRB1 A0A1S3AAW1 A0A1D2A4H9 B4I0A1 A0A182VNH7 A0A3Q1FE22 A0A3P9H7B2 A0A3P9KGC4 I3KT22 A0A3B4VM22 A0A3Q3QX64 Q3LSM4 A0A1Y1KT54 A0A2A3ERY3 K7IQ85 A0A182H713 Q16FD8 A0A3P8VG94 A0A0M4F8F0 N6USA5 A0A182KX75 H2MYK7 A0A3Q4HB18 A0A182M4C6 Q7QH32 A0A3P9KFX5 B0WZW4 A0A3P9H7C5 A0A182WW14 A0A182R878 A0A1Q3FN94 A0A0L0BXN4 A0A3Q3GT65 A0A034VZ82 A0A0C9QS08 A0A0L7QXH0 B4JKK7 A0A154P0L2 A0A0K8W8D1 A0A0A1XJC1 A0A182FKZ2 A0A3B4WR59 A0A182HJY8 A0A3Q0RHC5 A0A182U6U7 A0A182YLN1 A0A182IPW9 A0A059LSR7 A0A2U9C6U5 A0A3Q1AJZ8 A0A1W4X347 A0A3B3ZUX2 B4L6E9 A0A088A573 A0A3R7NTN3 A0A182P8W5 A0A3Q1JI51 B4M824 A0A2K6EMJ8 Q6DIW8
A0A3S2N685 A0A2H1VZD4 A0A194PTF0 A0A2H1W814 A0A194QT08 A0A194PNT9 A0A0L7LVG8 D6WZD5 A0A1D2M7D5 A0A2J7QYG5 B4MTD7 U5EHZ8 A0A0M9ABU5 A0A0P4VW80 A0A0P4W285 A0A182NL11 V5IAQ9 B3MZ47 A0A1W4VFD6 Q9W3Z3 C6TP18 B3NXS8 A0A3B0KL32 T1PAB8 B4PYT0 B4R5M1 A0A3Q4MTI7 A0A3Q2WPW6 Q29J54 B4GTB5 A0A3B4GVH1 A0A087SQK2 A0A3P8RB99 A0A3P9BRB1 A0A1S3AAW1 A0A1D2A4H9 B4I0A1 A0A182VNH7 A0A3Q1FE22 A0A3P9H7B2 A0A3P9KGC4 I3KT22 A0A3B4VM22 A0A3Q3QX64 Q3LSM4 A0A1Y1KT54 A0A2A3ERY3 K7IQ85 A0A182H713 Q16FD8 A0A3P8VG94 A0A0M4F8F0 N6USA5 A0A182KX75 H2MYK7 A0A3Q4HB18 A0A182M4C6 Q7QH32 A0A3P9KFX5 B0WZW4 A0A3P9H7C5 A0A182WW14 A0A182R878 A0A1Q3FN94 A0A0L0BXN4 A0A3Q3GT65 A0A034VZ82 A0A0C9QS08 A0A0L7QXH0 B4JKK7 A0A154P0L2 A0A0K8W8D1 A0A0A1XJC1 A0A182FKZ2 A0A3B4WR59 A0A182HJY8 A0A3Q0RHC5 A0A182U6U7 A0A182YLN1 A0A182IPW9 A0A059LSR7 A0A2U9C6U5 A0A3Q1AJZ8 A0A1W4X347 A0A3B3ZUX2 B4L6E9 A0A088A573 A0A3R7NTN3 A0A182P8W5 A0A3Q1JI51 B4M824 A0A2K6EMJ8 Q6DIW8
EC Number
2.6.1.44
2.6.1.51
2.6.1.51
Pubmed
19121390
28756777
22118469
26354079
26227816
18362917
+ More
19820115 27289101 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304 25186727 15632085 25012212 17554307 16681462 16990263 17510324 28004739 20075255 26483478 24487278 23537049 20966253 12364791 14747013 17210077 26108605 25348373 25830018 25244985 24809511 25463417
19820115 27289101 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304 25186727 15632085 25012212 17554307 16681462 16990263 17510324 28004739 20075255 26483478 24487278 23537049 20966253 12364791 14747013 17210077 26108605 25348373 25830018 25244985 24809511 25463417
EMBL
BABH01013820
BABH01013821
NWSH01002333
PCG68508.1
KZ149904
PZC78379.1
+ More
PZC78378.1 PCG68509.1 AGBW02010228 OWR48881.1 RSAL01000320 RVE42562.1 ODYU01005253 SOQ45922.1 KQ459599 KPI94410.1 ODYU01006802 SOQ48992.1 KQ461154 KPJ08454.1 KPI94409.1 JTDY01000051 KOB79161.1 KQ971372 EFA09724.1 LJIJ01003131 ODM88834.1 NEVH01009090 PNF33614.1 CH963851 EDW75376.1 GANO01002824 JAB57047.1 KQ435687 KOX81337.1 GDRN01109190 JAI57113.1 GDRN01109192 JAI57111.1 GALX01000755 JAB67711.1 CH902632 EDV32891.1 AE014298 BT133279 AAF46168.1 AFC38910.1 BT099504 ACU24744.1 CH954180 EDV47379.1 OUUW01000019 SPP89310.1 KA645721 AFP60350.1 CM000162 EDX01016.1 CM000366 EDX17258.1 CH379063 EAL32448.1 CH479190 EDW25785.1 KL662160 QOKY01000184 KFM28006.1 RMZ53954.1 GDKF01004538 JAT74084.1 CH480819 EDW52932.1 AERX01011518 DQ182329 CH477665 ABA26661.1 EAT37540.1 GEZM01077174 JAV63380.1 KZ288191 PBC34467.1 JXUM01115355 KQ565871 KXJ70586.1 CH478461 EAT32945.1 CP012528 ALC48637.1 APGK01018692 KB740082 ENN81587.1 AXCM01000231 AAAB01008820 EAA05410.3 DS232219 EDS37789.1 GFDL01005975 JAV29070.1 JRES01001179 KNC24756.1 GAKP01012084 JAC46868.1 GBYB01003422 JAG73189.1 KQ414705 KOC63256.1 CH916370 EDW00110.1 KQ434783 KZC04788.1 GDHF01004977 JAI47337.1 GBXI01017238 GBXI01002843 JAC97053.1 JAD11449.1 APCN01002223 AYPS01000026 KDD77074.1 CP026254 AWP11446.1 CH933812 EDW05945.1 QCYY01002941 ROT66404.1 CH940653 EDW62300.1 BC075417 BC089751 AAH75417.1 AAH89751.1
PZC78378.1 PCG68509.1 AGBW02010228 OWR48881.1 RSAL01000320 RVE42562.1 ODYU01005253 SOQ45922.1 KQ459599 KPI94410.1 ODYU01006802 SOQ48992.1 KQ461154 KPJ08454.1 KPI94409.1 JTDY01000051 KOB79161.1 KQ971372 EFA09724.1 LJIJ01003131 ODM88834.1 NEVH01009090 PNF33614.1 CH963851 EDW75376.1 GANO01002824 JAB57047.1 KQ435687 KOX81337.1 GDRN01109190 JAI57113.1 GDRN01109192 JAI57111.1 GALX01000755 JAB67711.1 CH902632 EDV32891.1 AE014298 BT133279 AAF46168.1 AFC38910.1 BT099504 ACU24744.1 CH954180 EDV47379.1 OUUW01000019 SPP89310.1 KA645721 AFP60350.1 CM000162 EDX01016.1 CM000366 EDX17258.1 CH379063 EAL32448.1 CH479190 EDW25785.1 KL662160 QOKY01000184 KFM28006.1 RMZ53954.1 GDKF01004538 JAT74084.1 CH480819 EDW52932.1 AERX01011518 DQ182329 CH477665 ABA26661.1 EAT37540.1 GEZM01077174 JAV63380.1 KZ288191 PBC34467.1 JXUM01115355 KQ565871 KXJ70586.1 CH478461 EAT32945.1 CP012528 ALC48637.1 APGK01018692 KB740082 ENN81587.1 AXCM01000231 AAAB01008820 EAA05410.3 DS232219 EDS37789.1 GFDL01005975 JAV29070.1 JRES01001179 KNC24756.1 GAKP01012084 JAC46868.1 GBYB01003422 JAG73189.1 KQ414705 KOC63256.1 CH916370 EDW00110.1 KQ434783 KZC04788.1 GDHF01004977 JAI47337.1 GBXI01017238 GBXI01002843 JAC97053.1 JAD11449.1 APCN01002223 AYPS01000026 KDD77074.1 CP026254 AWP11446.1 CH933812 EDW05945.1 QCYY01002941 ROT66404.1 CH940653 EDW62300.1 BC075417 BC089751 AAH75417.1 AAH89751.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
+ More
UP000037510 UP000007266 UP000094527 UP000235965 UP000007798 UP000053105 UP000075884 UP000007801 UP000192221 UP000000803 UP000008711 UP000268350 UP000095301 UP000002282 UP000000304 UP000261580 UP000264840 UP000001819 UP000008744 UP000261460 UP000028924 UP000279271 UP000265100 UP000265160 UP000079721 UP000001292 UP000075903 UP000257200 UP000265200 UP000265180 UP000005207 UP000261420 UP000261600 UP000008820 UP000242457 UP000002358 UP000069940 UP000249989 UP000265120 UP000092553 UP000019118 UP000075882 UP000001038 UP000075883 UP000007062 UP000002320 UP000076407 UP000075900 UP000037069 UP000261660 UP000053825 UP000001070 UP000076502 UP000069272 UP000261360 UP000075840 UP000261340 UP000075902 UP000076408 UP000075880 UP000026042 UP000246464 UP000257160 UP000192223 UP000261520 UP000009192 UP000005203 UP000283509 UP000075885 UP000265040 UP000008792 UP000233160
UP000037510 UP000007266 UP000094527 UP000235965 UP000007798 UP000053105 UP000075884 UP000007801 UP000192221 UP000000803 UP000008711 UP000268350 UP000095301 UP000002282 UP000000304 UP000261580 UP000264840 UP000001819 UP000008744 UP000261460 UP000028924 UP000279271 UP000265100 UP000265160 UP000079721 UP000001292 UP000075903 UP000257200 UP000265200 UP000265180 UP000005207 UP000261420 UP000261600 UP000008820 UP000242457 UP000002358 UP000069940 UP000249989 UP000265120 UP000092553 UP000019118 UP000075882 UP000001038 UP000075883 UP000007062 UP000002320 UP000076407 UP000075900 UP000037069 UP000261660 UP000053825 UP000001070 UP000076502 UP000069272 UP000261360 UP000075840 UP000261340 UP000075902 UP000076408 UP000075880 UP000026042 UP000246464 UP000257160 UP000192223 UP000261520 UP000009192 UP000005203 UP000283509 UP000075885 UP000265040 UP000008792 UP000233160
Pfam
PF00266 Aminotran_5
Interpro
SUPFAM
SSF53383
SSF53383
Gene 3D
ProteinModelPortal
H9J7G6
A0A2A4JA13
A0A2W1BTN4
A0A2W1BTI9
A0A2A4J9J0
A0A212F566
+ More
A0A3S2N685 A0A2H1VZD4 A0A194PTF0 A0A2H1W814 A0A194QT08 A0A194PNT9 A0A0L7LVG8 D6WZD5 A0A1D2M7D5 A0A2J7QYG5 B4MTD7 U5EHZ8 A0A0M9ABU5 A0A0P4VW80 A0A0P4W285 A0A182NL11 V5IAQ9 B3MZ47 A0A1W4VFD6 Q9W3Z3 C6TP18 B3NXS8 A0A3B0KL32 T1PAB8 B4PYT0 B4R5M1 A0A3Q4MTI7 A0A3Q2WPW6 Q29J54 B4GTB5 A0A3B4GVH1 A0A087SQK2 A0A3P8RB99 A0A3P9BRB1 A0A1S3AAW1 A0A1D2A4H9 B4I0A1 A0A182VNH7 A0A3Q1FE22 A0A3P9H7B2 A0A3P9KGC4 I3KT22 A0A3B4VM22 A0A3Q3QX64 Q3LSM4 A0A1Y1KT54 A0A2A3ERY3 K7IQ85 A0A182H713 Q16FD8 A0A3P8VG94 A0A0M4F8F0 N6USA5 A0A182KX75 H2MYK7 A0A3Q4HB18 A0A182M4C6 Q7QH32 A0A3P9KFX5 B0WZW4 A0A3P9H7C5 A0A182WW14 A0A182R878 A0A1Q3FN94 A0A0L0BXN4 A0A3Q3GT65 A0A034VZ82 A0A0C9QS08 A0A0L7QXH0 B4JKK7 A0A154P0L2 A0A0K8W8D1 A0A0A1XJC1 A0A182FKZ2 A0A3B4WR59 A0A182HJY8 A0A3Q0RHC5 A0A182U6U7 A0A182YLN1 A0A182IPW9 A0A059LSR7 A0A2U9C6U5 A0A3Q1AJZ8 A0A1W4X347 A0A3B3ZUX2 B4L6E9 A0A088A573 A0A3R7NTN3 A0A182P8W5 A0A3Q1JI51 B4M824 A0A2K6EMJ8 Q6DIW8
A0A3S2N685 A0A2H1VZD4 A0A194PTF0 A0A2H1W814 A0A194QT08 A0A194PNT9 A0A0L7LVG8 D6WZD5 A0A1D2M7D5 A0A2J7QYG5 B4MTD7 U5EHZ8 A0A0M9ABU5 A0A0P4VW80 A0A0P4W285 A0A182NL11 V5IAQ9 B3MZ47 A0A1W4VFD6 Q9W3Z3 C6TP18 B3NXS8 A0A3B0KL32 T1PAB8 B4PYT0 B4R5M1 A0A3Q4MTI7 A0A3Q2WPW6 Q29J54 B4GTB5 A0A3B4GVH1 A0A087SQK2 A0A3P8RB99 A0A3P9BRB1 A0A1S3AAW1 A0A1D2A4H9 B4I0A1 A0A182VNH7 A0A3Q1FE22 A0A3P9H7B2 A0A3P9KGC4 I3KT22 A0A3B4VM22 A0A3Q3QX64 Q3LSM4 A0A1Y1KT54 A0A2A3ERY3 K7IQ85 A0A182H713 Q16FD8 A0A3P8VG94 A0A0M4F8F0 N6USA5 A0A182KX75 H2MYK7 A0A3Q4HB18 A0A182M4C6 Q7QH32 A0A3P9KFX5 B0WZW4 A0A3P9H7C5 A0A182WW14 A0A182R878 A0A1Q3FN94 A0A0L0BXN4 A0A3Q3GT65 A0A034VZ82 A0A0C9QS08 A0A0L7QXH0 B4JKK7 A0A154P0L2 A0A0K8W8D1 A0A0A1XJC1 A0A182FKZ2 A0A3B4WR59 A0A182HJY8 A0A3Q0RHC5 A0A182U6U7 A0A182YLN1 A0A182IPW9 A0A059LSR7 A0A2U9C6U5 A0A3Q1AJZ8 A0A1W4X347 A0A3B3ZUX2 B4L6E9 A0A088A573 A0A3R7NTN3 A0A182P8W5 A0A3Q1JI51 B4M824 A0A2K6EMJ8 Q6DIW8
PDB
2HUU
E-value=1.14902e-76,
Score=729
Ontologies
PATHWAY
00250
Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00260 Glycine, serine and threonine metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
04146 Peroxisome - Bombyx mori (domestic silkworm)
00260 Glycine, serine and threonine metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
04146 Peroxisome - Bombyx mori (domestic silkworm)
GO
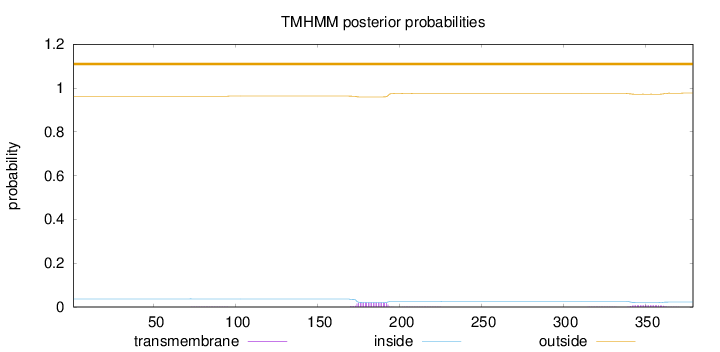
Topology
Length:
379
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.574530000000001
Exp number, first 60 AAs:
1e-05
Total prob of N-in:
0.03708
outside
1 - 379
Population Genetic Test Statistics
Pi
197.737272
Theta
176.370234
Tajima's D
-0.881838
CLR
181.20286
CSRT
0.15859207039648
Interpretation
Uncertain
