Gene
KWMTBOMO04519
Pre Gene Modal
BGIBMGA005352
Annotation
PREDICTED:_transducin_beta-like_protein_2_isoform_X2_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 2.894
Sequence
CDS
ATGGGAGAGATTGAAAGCGGTCAAAACATTCACGTGTTCCTCTTGTCGGCAACTGCAGTTTTAGTCTGCGTTTTGTTGCATTTCATTTACAAAATTATATTCCGCAACAAAAACCAAGCGGTGGAGGAACGTGAGAGAGAGAACAATTCTGGCACTCTCACTGATGAACCGCCGTGTGAAGCCGTTGCGGAGACACGCGTCGGCGGTAAAAGCAAAAAACGCCCAGCATGGAAAGGAAAAACGGACTTCAGCCATCCCTGGCTTATGAAGAATCTCAAGGGTCATCCCGGAACGGTTTTGTTAATGGATTTCTCCGCCAACGGCAAGTTCATGGCCGCTACCTGTGATGGTAAGTTCTCTCCAACATTTACAAATGCTTAA
Protein
MGEIESGQNIHVFLLSATAVLVCVLLHFIYKIIFRNKNQAVEERERENNSGTLTDEPPCEAVAETRVGGKSKKRPAWKGKTDFSHPWLMKNLKGHPGTVLLMDFSANGKFMAATCDGKFSPTFTNA
Summary
Uniprot
H9J760
A0A2H1WJN3
A0A2W1BTH2
A0A194PTF5
S4Q0C7
A0A194QSI4
+ More
A0A212F6P6 A0A1E1WEL1 A0A1E1WND9 A0A0A9Y2A4 B7QL80 A0A146MB66 A0A147BEV3 A0A1Y1MZI9 A0A1Y1N1I3 R4G3I1 K1Q541 A0A139WCZ4 A0A0C9PMH3 E2BXL5 U5EHC5 F7ATG5 A0A1W2W4K0 A0A0R3QW65 A0A0C2CS42 A0A2L2XVY6 A0A146QZ75 A0A183GXB2 A0A3P8EF96 A0A146UZU6 A0A3Q2QZI4 A0A3L8DHF0 A0A146V0A5 A0A146UZT9 A0A182ECV2 A0A044QND0 A0A1S0UJB3 A0A146MLX8 A0A1B6DK88 A0A0K0JDD1 A0A1S3RLC9 A0A0I9N6A8 A0A3S1C787 A0A3B3T0F3 A0A1S3LQ99 A0A0L7R5I1 A0A0N4YQG0
A0A212F6P6 A0A1E1WEL1 A0A1E1WND9 A0A0A9Y2A4 B7QL80 A0A146MB66 A0A147BEV3 A0A1Y1MZI9 A0A1Y1N1I3 R4G3I1 K1Q541 A0A139WCZ4 A0A0C9PMH3 E2BXL5 U5EHC5 F7ATG5 A0A1W2W4K0 A0A0R3QW65 A0A0C2CS42 A0A2L2XVY6 A0A146QZ75 A0A183GXB2 A0A3P8EF96 A0A146UZU6 A0A3Q2QZI4 A0A3L8DHF0 A0A146V0A5 A0A146UZT9 A0A182ECV2 A0A044QND0 A0A1S0UJB3 A0A146MLX8 A0A1B6DK88 A0A0K0JDD1 A0A1S3RLC9 A0A0I9N6A8 A0A3S1C787 A0A3B3T0F3 A0A1S3LQ99 A0A0L7R5I1 A0A0N4YQG0
Pubmed
EMBL
BABH01013792
ODYU01009024
SOQ53122.1
KZ149904
PZC78372.1
KQ459599
+ More
KPI94415.1 GAIX01000018 JAA92542.1 KQ461154 KPJ08447.1 AGBW02009987 OWR49398.1 GDQN01005631 JAT85423.1 GDQN01002511 JAT88543.1 GBHO01045019 GBHO01045018 GBHO01019979 GBHO01019783 GBRD01003670 JAF98584.1 JAF98585.1 JAG23625.1 JAG23821.1 JAG62151.1 ABJB010034310 ABJB010725763 ABJB010959403 DS963933 EEC19602.1 GDHC01004172 GDHC01001611 JAQ14457.1 JAQ17018.1 GEGO01006064 JAR89340.1 GEZM01020113 JAV89396.1 GEZM01020112 JAV89397.1 ACPB03015393 GAHY01002185 JAA75325.1 JH816849 EKC28998.1 KQ971361 KYB25819.1 GBYB01002273 JAG72040.1 GL451265 EFN79584.1 GANO01003084 JAB56787.1 EAAA01000293 UZAG01017289 VDO33941.1 KN731728 KIH59653.1 IAAA01003393 IAAA01003394 LAA00256.1 GCES01124899 JAQ61423.1 UZAH01043088 VDP62787.1 GCES01075597 JAR10726.1 QOIP01000008 RLU19771.1 GCES01075598 JAR10725.1 GCES01075599 JAR10724.1 UYRW01001696 VDK80007.1 CMVM020000328 JH712107 EJD75802.1 GCES01165621 JAQ20701.1 GEDC01011268 JAS26030.1 LN857020 CTP81521.1 RQTK01000192 RUS84826.1 KQ414652 KOC66103.1 UYSL01024218 VDL83217.1
KPI94415.1 GAIX01000018 JAA92542.1 KQ461154 KPJ08447.1 AGBW02009987 OWR49398.1 GDQN01005631 JAT85423.1 GDQN01002511 JAT88543.1 GBHO01045019 GBHO01045018 GBHO01019979 GBHO01019783 GBRD01003670 JAF98584.1 JAF98585.1 JAG23625.1 JAG23821.1 JAG62151.1 ABJB010034310 ABJB010725763 ABJB010959403 DS963933 EEC19602.1 GDHC01004172 GDHC01001611 JAQ14457.1 JAQ17018.1 GEGO01006064 JAR89340.1 GEZM01020113 JAV89396.1 GEZM01020112 JAV89397.1 ACPB03015393 GAHY01002185 JAA75325.1 JH816849 EKC28998.1 KQ971361 KYB25819.1 GBYB01002273 JAG72040.1 GL451265 EFN79584.1 GANO01003084 JAB56787.1 EAAA01000293 UZAG01017289 VDO33941.1 KN731728 KIH59653.1 IAAA01003393 IAAA01003394 LAA00256.1 GCES01124899 JAQ61423.1 UZAH01043088 VDP62787.1 GCES01075597 JAR10726.1 QOIP01000008 RLU19771.1 GCES01075598 JAR10725.1 GCES01075599 JAR10724.1 UYRW01001696 VDK80007.1 CMVM020000328 JH712107 EJD75802.1 GCES01165621 JAQ20701.1 GEDC01011268 JAS26030.1 LN857020 CTP81521.1 RQTK01000192 RUS84826.1 KQ414652 KOC66103.1 UYSL01024218 VDL83217.1
Proteomes
PRIDE
Interpro
IPR019775
WD40_repeat_CS
+ More
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR012337 RNaseH-like_sf
IPR034922 REX1-like_exo
IPR013520 Exonuclease_RNaseT/DNA_pol3
IPR036397 RNaseH_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR020472 G-protein_beta_WD-40_rep
IPR031736 EloA-BP1
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR012337 RNaseH-like_sf
IPR034922 REX1-like_exo
IPR013520 Exonuclease_RNaseT/DNA_pol3
IPR036397 RNaseH_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR020472 G-protein_beta_WD-40_rep
IPR031736 EloA-BP1
Gene 3D
ProteinModelPortal
H9J760
A0A2H1WJN3
A0A2W1BTH2
A0A194PTF5
S4Q0C7
A0A194QSI4
+ More
A0A212F6P6 A0A1E1WEL1 A0A1E1WND9 A0A0A9Y2A4 B7QL80 A0A146MB66 A0A147BEV3 A0A1Y1MZI9 A0A1Y1N1I3 R4G3I1 K1Q541 A0A139WCZ4 A0A0C9PMH3 E2BXL5 U5EHC5 F7ATG5 A0A1W2W4K0 A0A0R3QW65 A0A0C2CS42 A0A2L2XVY6 A0A146QZ75 A0A183GXB2 A0A3P8EF96 A0A146UZU6 A0A3Q2QZI4 A0A3L8DHF0 A0A146V0A5 A0A146UZT9 A0A182ECV2 A0A044QND0 A0A1S0UJB3 A0A146MLX8 A0A1B6DK88 A0A0K0JDD1 A0A1S3RLC9 A0A0I9N6A8 A0A3S1C787 A0A3B3T0F3 A0A1S3LQ99 A0A0L7R5I1 A0A0N4YQG0
A0A212F6P6 A0A1E1WEL1 A0A1E1WND9 A0A0A9Y2A4 B7QL80 A0A146MB66 A0A147BEV3 A0A1Y1MZI9 A0A1Y1N1I3 R4G3I1 K1Q541 A0A139WCZ4 A0A0C9PMH3 E2BXL5 U5EHC5 F7ATG5 A0A1W2W4K0 A0A0R3QW65 A0A0C2CS42 A0A2L2XVY6 A0A146QZ75 A0A183GXB2 A0A3P8EF96 A0A146UZU6 A0A3Q2QZI4 A0A3L8DHF0 A0A146V0A5 A0A146UZT9 A0A182ECV2 A0A044QND0 A0A1S0UJB3 A0A146MLX8 A0A1B6DK88 A0A0K0JDD1 A0A1S3RLC9 A0A0I9N6A8 A0A3S1C787 A0A3B3T0F3 A0A1S3LQ99 A0A0L7R5I1 A0A0N4YQG0
Ontologies
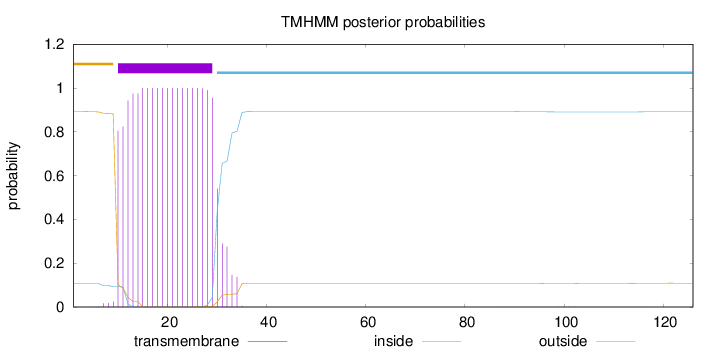
Topology
Length:
126
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.94402
Exp number, first 60 AAs:
20.91853
Total prob of N-in:
0.10761
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 29
inside
30 - 126
Population Genetic Test Statistics
Pi
250.191053
Theta
188.154608
Tajima's D
1.121347
CLR
0.002774
CSRT
0.696665166741663
Interpretation
Uncertain
