Gene
KWMTBOMO04517
Pre Gene Modal
BGIBMGA005454
Annotation
PREDICTED:_tripartite_motif-containing_protein_2-like_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.135
Sequence
CDS
ATGTATTGTTGTTCGTTTCTCTCACGGCTGGGTGTCGGGATGGCGAGCATGAGTTCCACGCTCGTCGAAACCGTCTCCATCAACTACGAGGACTTTAATGAAAGTTTCCTCACATGCGGCACATGTTTATGCACTTATGATGGAGGCGAACACACCCCGAAGCTTCTGCCGTGCTCACACACGGTTTGTCTTCATTGCCTAACGCGCATCGCGGCTTCACAGACACGGGACGCCGGCAGCTTTCGATGTCCGATTTGTCGCGAACTGATCACAATACCTCGAGGAGGGGTACCGGCTCTCCCGCCTTCGTTTCTCGTCAATCAGCTGCTCGATCTTATGGCACGGCAACGCCGAGAGGTTATACCGCAGTGTAGCTCGCACCCAGGCAGGGAGCTACTTTTCTGCGAGACATGCGATTGCGTTTTTTGTCGTTACTGTGCCGATGGACCGCACAGCGACACGCCGTGTGACCACACAGTTGTACCGTTTTCGATAGCGTTGAAAAGGATGTCAGAAATATTGCTATACAGAGCCAACGAGTGCCTGAGCAAATTAGGTTCGGCTAGGGAAGCGGTAGCAGGTGAATTGAGGAGATTGGAAGCGGCTGCAACTGCGGCAGACGAAGCTATTGATAGACACTTTGCGGAACTGAGAGCGGCACTAGAGAAGAGGCATAATGAATTAAAGTCTGCCGCCGCAGCGGCTGCCAGTCACAAACGAAAGTTACTCGAAGAACAATTGAAGCTGATTGATGCAGAGAAATCTAAAGTGGAAGCTGAGTGTTCCGGGTTACAGCAACAGGTAGAAGTGCGTGAAGTGAGCAGCAGAGCGGCTGCTTTGGGCGCCAGATTGGGTGACGCGGCTGCATTGGCCGAGCCAAGAGAGAACGCGTTCTTAGCCGTCGACTTCGCGCACAACGACGCTCAACAACGCTTCGTGGATTCTCTCGAAGTGCTCGGTCGTGTCAGGACCAGTACTACTTTCCCAGGGCTGTGTTCGTTACAACTCGAATCATCATGCGTGTCTGGTTTGGAAGTGGTGGTGGTTCTCCGCACCGTGGACTATCACGGTGAGGCGCGTAGCACCGGCGGGGACCCCGTGACGGCGGCCGCCACGCTCGACGACGCCCCTCTGCCTTGCACCGTCACCGATCTGGATTCCGGACTCTATAGGATCTCGATGCGTCCGTGGAGTGCGGGCTCGGTGTCGGTGCGCGTGCTGGTGTTCTCCCGCGCCGTGCGCGACTCGCCGCTGCGCGCCGCCGTGACGTCACAAGCCGCGCCGCTGCGGGTGTGGGGCGCGCGGGGGAACGGGAAGGACCAGCTGAGCCAGCCCGTCGCGCTAGCCAGATGTCCGAAGCGTAAAGAAATTTATGTTCTGGATGCTGGTAACTCAAGAGTGAAAGTGTTGGACGACGAGACGTTCGCTTTCAAAACGCATATCGTGAATGAAGGACTGGCGGGTCGCAGTTGTACCGGCATTGCTGTGACCGCGGAGGGCGTGGCCGCCGTCAATTGGAGGACTCGCACTATCACGGAGGTGAACACAGAAGGTACGACGCTAAGAACGATTCGTTCGGATCGTCTGGTGGAGCCGGTGTGCGTGGCGGCCGACGTGGCGACGCGACGTCTGGCCGTCGCCGACAACGGCGCCAGAACTGTCTTCGTCTTCGATCGCTACGGAAACATCGAAAGAGAAATAGGCAACGGTGATCTCGGCTTAGTGGGTGGTCTAGCTTTATCCGAAGACCTCATCATCATAGCTGACGTGGCGGTGCGGATCTACGATTCTGAAGGAAATCTCAAAAACACATTGGCCCCCGTCCCCAAAGGTCGAGGGGTGTACGGTGGGGTGGCTATAGACGAGACCGGCAGCGGCAAGCACATCGTGTGCGCTCGCTCGGTGCGCGGGCGGGCCGCCCTCGTGGTGCTGGAGGCGGGGGGCGGCGGGAAGACGCTGGCGGCCTGCGAGCTCCTCGACAAGAAGCCGCGGAGGGTCGCCGGCATCGCGCTGCTGCCCAACAAGAAGGTGCTGCTAGCCGACCTGGCGGGAGACTGCTTAAGACTGCACACTTACTGGTGA
Protein
MYCCSFLSRLGVGMASMSSTLVETVSINYEDFNESFLTCGTCLCTYDGGEHTPKLLPCSHTVCLHCLTRIAASQTRDAGSFRCPICRELITIPRGGVPALPPSFLVNQLLDLMARQRREVIPQCSSHPGRELLFCETCDCVFCRYCADGPHSDTPCDHTVVPFSIALKRMSEILLYRANECLSKLGSAREAVAGELRRLEAAATAADEAIDRHFAELRAALEKRHNELKSAAAAAASHKRKLLEEQLKLIDAEKSKVEAECSGLQQQVEVREVSSRAAALGARLGDAAALAEPRENAFLAVDFAHNDAQQRFVDSLEVLGRVRTSTTFPGLCSLQLESSCVSGLEVVVVLRTVDYHGEARSTGGDPVTAAATLDDAPLPCTVTDLDSGLYRISMRPWSAGSVSVRVLVFSRAVRDSPLRAAVTSQAAPLRVWGARGNGKDQLSQPVALARCPKRKEIYVLDAGNSRVKVLDDETFAFKTHIVNEGLAGRSCTGIAVTAEGVAAVNWRTRTITEVNTEGTTLRTIRSDRLVEPVCVAADVATRRLAVADNGARTVFVFDRYGNIEREIGNGDLGLVGGLALSEDLIIIADVAVRIYDSEGNLKNTLAPVPKGRGVYGGVAIDETGSGKHIVCARSVRGRAALVVLEAGGGGKTLAACELLDKKPRRVAGIALLPNKKVLLADLAGDCLRLHTYW
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9J7G2
A0A2H1WJ77
A0A2W1BVK2
A0A212EU18
A0A194PN49
A0A194QSE3
+ More
A0A0L7LAU3 A0A151IT52 A0A1Y1MW86 D6X1L3 A0A151IHA4 A0A087ZQ86 A0A1Y1MXK3 A0A195BX43 A0A3L8DI78 A0A2J7PTV3 A0A195F165 A0A026WT67 K7IRM5 A0A139WCH6 E2ACM2 A0A139WCL8 A0A067RB97 A0A1W4W8A5 A0A1W4WJK0 E2BNV4 A0A232EYW1 A0A2A3EIX6 A0A310STZ2 A0A139WCH5 A0A1B6EPD4 E0W0L9 A0A154P438 A0A1W4WIK8 A0A1W4WJU5 A0A0M9A2G0 A0A2J7PTV5 A0A151XG02 A0A0L7R4B5 A0A1Y1MZN3 A0A1Y1MW11 A0A1W4W8A3 A0A0C9RGY2 A0A0A9XTU7 A0A146M342 A0A1S3HIE6 A0A0P4VY24 V4A8J1 A0A3S5WGY9 A0A3S3NXN4 A0A210Q5K8 A0A0P4VT58 A0A023F0M5 A0A2P6KLZ8 A0A0P4W0C3 A0A2P8YRY4 A0A2C9K4L2 R7V8F5 T1IJA1 A0A0B7A9T4 A0A0L8FG06 A0A0K2V557 A0A0T6AWK9 T1ELW1 T1KXQ6 T1KXG2 A0A147BPZ7 A0A1Y1MW26 T1IBP8 L7MDZ2 A0A131YFM6 A0A1E1XHI1 F4WAU9 A0A132AD25 A0A0V1GZU4 A0A0V1MKU9 A0A1D1V988 A0A0V0V9N3 A0A2T7PQ59 F1KUD7 A0A0J7KPV1 A0A1Y3E554 A0A0V1G180 A0A0V1DZK2 A0A0V1IER3 A0A0V0S1G7 A0A0V0ZRI6 A0A183UHW9 A0A0N4UAR5 A0A267GZ12 A0A0V1CIL3 A0A0B2VYF2 A0A0V1CHV4 A0A0V1KZ58 A0A0V1L001 A0A0V0VA23 A0A0V1NTJ2
A0A0L7LAU3 A0A151IT52 A0A1Y1MW86 D6X1L3 A0A151IHA4 A0A087ZQ86 A0A1Y1MXK3 A0A195BX43 A0A3L8DI78 A0A2J7PTV3 A0A195F165 A0A026WT67 K7IRM5 A0A139WCH6 E2ACM2 A0A139WCL8 A0A067RB97 A0A1W4W8A5 A0A1W4WJK0 E2BNV4 A0A232EYW1 A0A2A3EIX6 A0A310STZ2 A0A139WCH5 A0A1B6EPD4 E0W0L9 A0A154P438 A0A1W4WIK8 A0A1W4WJU5 A0A0M9A2G0 A0A2J7PTV5 A0A151XG02 A0A0L7R4B5 A0A1Y1MZN3 A0A1Y1MW11 A0A1W4W8A3 A0A0C9RGY2 A0A0A9XTU7 A0A146M342 A0A1S3HIE6 A0A0P4VY24 V4A8J1 A0A3S5WGY9 A0A3S3NXN4 A0A210Q5K8 A0A0P4VT58 A0A023F0M5 A0A2P6KLZ8 A0A0P4W0C3 A0A2P8YRY4 A0A2C9K4L2 R7V8F5 T1IJA1 A0A0B7A9T4 A0A0L8FG06 A0A0K2V557 A0A0T6AWK9 T1ELW1 T1KXQ6 T1KXG2 A0A147BPZ7 A0A1Y1MW26 T1IBP8 L7MDZ2 A0A131YFM6 A0A1E1XHI1 F4WAU9 A0A132AD25 A0A0V1GZU4 A0A0V1MKU9 A0A1D1V988 A0A0V0V9N3 A0A2T7PQ59 F1KUD7 A0A0J7KPV1 A0A1Y3E554 A0A0V1G180 A0A0V1DZK2 A0A0V1IER3 A0A0V0S1G7 A0A0V0ZRI6 A0A183UHW9 A0A0N4UAR5 A0A267GZ12 A0A0V1CIL3 A0A0B2VYF2 A0A0V1CHV4 A0A0V1KZ58 A0A0V1L001 A0A0V0VA23 A0A0V1NTJ2
Pubmed
EMBL
BABH01013789
ODYU01009024
SOQ53125.1
KZ149904
PZC78371.1
AGBW02012491
+ More
OWR44986.1 KQ459599 KPI94418.1 KQ461154 KPJ08443.1 JTDY01001897 KOB72597.1 KQ981039 KYN10143.1 GEZM01019125 JAV89863.1 KQ971370 EFA09351.2 KQ977636 KYN01156.1 GEZM01019131 JAV89858.1 KQ976394 KYM93147.1 QOIP01000008 RLU19902.1 NEVH01021221 PNF19762.1 KQ981880 KYN33912.1 KK107109 EZA59133.1 KYB25604.1 GL438568 EFN68766.1 KYB25605.1 KK852619 KDR20098.1 GL449507 EFN82637.1 NNAY01001573 OXU23551.1 KZ288229 PBC31725.1 KQ759887 OAD62128.1 KYB25606.1 GECZ01029988 JAS39781.1 AAZO01006794 DS235861 EEB19175.1 KQ434809 KZC06603.1 KQ435756 KOX75915.1 PNF19763.1 KQ982174 KYQ59314.1 KQ414658 KOC65664.1 GEZM01019137 JAV89855.1 GEZM01019127 JAV89862.1 GBYB01007585 JAG77352.1 GBHO01038147 GBHO01027782 GBHO01020271 GBHO01020270 GBHO01020267 JAG05457.1 JAG15822.1 JAG23333.1 JAG23334.1 JAG23337.1 GDHC01005217 JAQ13412.1 GDRN01099147 JAI58826.1 KB202283 ESO91335.1 NCKU01002966 RWS08444.1 NCKU01003456 NCKU01003187 RWS07481.1 RWS08004.1 NEDP02004925 OWF44017.1 GDRN01099146 JAI58827.1 GBBI01004176 JAC14536.1 MWRG01008973 PRD27346.1 GDRN01099149 JAI58825.1 PYGN01000400 PSN47003.1 AMQN01005353 KB295796 ELU12631.1 JH430236 HACG01030897 HACG01030898 HACG01030899 CEK77762.1 CEK77763.1 CEK77764.1 KQ433008 KOF62572.1 HACA01028069 CDW45430.1 LJIG01022735 KRT78983.1 AMQM01002920 KB095959 ESO09257.1 CAEY01000696 CAEY01000695 GEGO01002959 JAR92445.1 GEZM01019128 JAV89861.1 ACPB03018351 GACK01002844 JAA62190.1 GEDV01010394 JAP78163.1 GFAC01000408 JAT98780.1 GL888053 EGI68705.1 JXLN01012343 KPM08360.1 JYDP01000181 KRZ03806.1 JYDO01000079 KRZ72429.1 BDGG01000004 GAU98239.1 JYDN01000062 KRX60226.1 PZQS01000002 PVD35561.1 JI166027 ADY41491.1 LBMM01004589 KMQ92264.1 LVZM01023116 OUC40253.1 JYDT01000009 KRY92011.1 JYDR01000153 KRY66916.1 JYDS01000211 JYDV01000093 KRZ21278.1 KRZ35262.1 JYDL01000048 KRX20537.1 JYDQ01000100 KRY15176.1 UYWY01019821 VDM39410.1 UYYG01001166 VDN58168.1 NIVC01000092 PAA91266.1 JYDI01000195 KRY48828.1 JPKZ01000619 KHN86459.1 KRY48827.1 JYDW01000188 KRZ52595.1 KRZ52594.1 KRX60225.1 JYDM01000102 KRZ87303.1
OWR44986.1 KQ459599 KPI94418.1 KQ461154 KPJ08443.1 JTDY01001897 KOB72597.1 KQ981039 KYN10143.1 GEZM01019125 JAV89863.1 KQ971370 EFA09351.2 KQ977636 KYN01156.1 GEZM01019131 JAV89858.1 KQ976394 KYM93147.1 QOIP01000008 RLU19902.1 NEVH01021221 PNF19762.1 KQ981880 KYN33912.1 KK107109 EZA59133.1 KYB25604.1 GL438568 EFN68766.1 KYB25605.1 KK852619 KDR20098.1 GL449507 EFN82637.1 NNAY01001573 OXU23551.1 KZ288229 PBC31725.1 KQ759887 OAD62128.1 KYB25606.1 GECZ01029988 JAS39781.1 AAZO01006794 DS235861 EEB19175.1 KQ434809 KZC06603.1 KQ435756 KOX75915.1 PNF19763.1 KQ982174 KYQ59314.1 KQ414658 KOC65664.1 GEZM01019137 JAV89855.1 GEZM01019127 JAV89862.1 GBYB01007585 JAG77352.1 GBHO01038147 GBHO01027782 GBHO01020271 GBHO01020270 GBHO01020267 JAG05457.1 JAG15822.1 JAG23333.1 JAG23334.1 JAG23337.1 GDHC01005217 JAQ13412.1 GDRN01099147 JAI58826.1 KB202283 ESO91335.1 NCKU01002966 RWS08444.1 NCKU01003456 NCKU01003187 RWS07481.1 RWS08004.1 NEDP02004925 OWF44017.1 GDRN01099146 JAI58827.1 GBBI01004176 JAC14536.1 MWRG01008973 PRD27346.1 GDRN01099149 JAI58825.1 PYGN01000400 PSN47003.1 AMQN01005353 KB295796 ELU12631.1 JH430236 HACG01030897 HACG01030898 HACG01030899 CEK77762.1 CEK77763.1 CEK77764.1 KQ433008 KOF62572.1 HACA01028069 CDW45430.1 LJIG01022735 KRT78983.1 AMQM01002920 KB095959 ESO09257.1 CAEY01000696 CAEY01000695 GEGO01002959 JAR92445.1 GEZM01019128 JAV89861.1 ACPB03018351 GACK01002844 JAA62190.1 GEDV01010394 JAP78163.1 GFAC01000408 JAT98780.1 GL888053 EGI68705.1 JXLN01012343 KPM08360.1 JYDP01000181 KRZ03806.1 JYDO01000079 KRZ72429.1 BDGG01000004 GAU98239.1 JYDN01000062 KRX60226.1 PZQS01000002 PVD35561.1 JI166027 ADY41491.1 LBMM01004589 KMQ92264.1 LVZM01023116 OUC40253.1 JYDT01000009 KRY92011.1 JYDR01000153 KRY66916.1 JYDS01000211 JYDV01000093 KRZ21278.1 KRZ35262.1 JYDL01000048 KRX20537.1 JYDQ01000100 KRY15176.1 UYWY01019821 VDM39410.1 UYYG01001166 VDN58168.1 NIVC01000092 PAA91266.1 JYDI01000195 KRY48828.1 JPKZ01000619 KHN86459.1 KRY48827.1 JYDW01000188 KRZ52595.1 KRZ52594.1 KRX60225.1 JYDM01000102 KRZ87303.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000037510
UP000078492
+ More
UP000007266 UP000078542 UP000005203 UP000078540 UP000279307 UP000235965 UP000078541 UP000053097 UP000002358 UP000000311 UP000027135 UP000192223 UP000008237 UP000215335 UP000242457 UP000009046 UP000076502 UP000053105 UP000075809 UP000053825 UP000085678 UP000030746 UP000285301 UP000242188 UP000245037 UP000076420 UP000014760 UP000053454 UP000015101 UP000015104 UP000015103 UP000007755 UP000055024 UP000054843 UP000186922 UP000054681 UP000245119 UP000036403 UP000243006 UP000054995 UP000054632 UP000054805 UP000054826 UP000054630 UP000054783 UP000050794 UP000267007 UP000038040 UP000274756 UP000215902 UP000054653 UP000031036 UP000054721 UP000054924
UP000007266 UP000078542 UP000005203 UP000078540 UP000279307 UP000235965 UP000078541 UP000053097 UP000002358 UP000000311 UP000027135 UP000192223 UP000008237 UP000215335 UP000242457 UP000009046 UP000076502 UP000053105 UP000075809 UP000053825 UP000085678 UP000030746 UP000285301 UP000242188 UP000245037 UP000076420 UP000014760 UP000053454 UP000015101 UP000015104 UP000015103 UP000007755 UP000055024 UP000054843 UP000186922 UP000054681 UP000245119 UP000036403 UP000243006 UP000054995 UP000054632 UP000054805 UP000054826 UP000054630 UP000054783 UP000050794 UP000267007 UP000038040 UP000274756 UP000215902 UP000054653 UP000031036 UP000054721 UP000054924
Pfam
Interpro
IPR011042
6-blade_b-propeller_TolB-like
+ More
IPR013083 Znf_RING/FYVE/PHD
IPR017868 Filamin/ABP280_repeat-like
IPR017907 Znf_RING_CS
IPR014756 Ig_E-set
IPR001841 Znf_RING
IPR000315 Znf_B-box
IPR013783 Ig-like_fold
IPR013017 NHL_repeat_subgr
IPR001298 Filamin/ABP280_rpt
IPR027370 Znf-RING_LisH
IPR001258 NHL_repeat
IPR026642 Glcci1/FAM117
IPR033116 TRYPSIN_SER
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR011011 Znf_FYVE_PHD
IPR018957 Znf_C3HC4_RING-type
IPR006954 MLT-10-like
IPR013083 Znf_RING/FYVE/PHD
IPR017868 Filamin/ABP280_repeat-like
IPR017907 Znf_RING_CS
IPR014756 Ig_E-set
IPR001841 Znf_RING
IPR000315 Znf_B-box
IPR013783 Ig-like_fold
IPR013017 NHL_repeat_subgr
IPR001298 Filamin/ABP280_rpt
IPR027370 Znf-RING_LisH
IPR001258 NHL_repeat
IPR026642 Glcci1/FAM117
IPR033116 TRYPSIN_SER
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR011011 Znf_FYVE_PHD
IPR018957 Znf_C3HC4_RING-type
IPR006954 MLT-10-like
Gene 3D
ProteinModelPortal
H9J7G2
A0A2H1WJ77
A0A2W1BVK2
A0A212EU18
A0A194PN49
A0A194QSE3
+ More
A0A0L7LAU3 A0A151IT52 A0A1Y1MW86 D6X1L3 A0A151IHA4 A0A087ZQ86 A0A1Y1MXK3 A0A195BX43 A0A3L8DI78 A0A2J7PTV3 A0A195F165 A0A026WT67 K7IRM5 A0A139WCH6 E2ACM2 A0A139WCL8 A0A067RB97 A0A1W4W8A5 A0A1W4WJK0 E2BNV4 A0A232EYW1 A0A2A3EIX6 A0A310STZ2 A0A139WCH5 A0A1B6EPD4 E0W0L9 A0A154P438 A0A1W4WIK8 A0A1W4WJU5 A0A0M9A2G0 A0A2J7PTV5 A0A151XG02 A0A0L7R4B5 A0A1Y1MZN3 A0A1Y1MW11 A0A1W4W8A3 A0A0C9RGY2 A0A0A9XTU7 A0A146M342 A0A1S3HIE6 A0A0P4VY24 V4A8J1 A0A3S5WGY9 A0A3S3NXN4 A0A210Q5K8 A0A0P4VT58 A0A023F0M5 A0A2P6KLZ8 A0A0P4W0C3 A0A2P8YRY4 A0A2C9K4L2 R7V8F5 T1IJA1 A0A0B7A9T4 A0A0L8FG06 A0A0K2V557 A0A0T6AWK9 T1ELW1 T1KXQ6 T1KXG2 A0A147BPZ7 A0A1Y1MW26 T1IBP8 L7MDZ2 A0A131YFM6 A0A1E1XHI1 F4WAU9 A0A132AD25 A0A0V1GZU4 A0A0V1MKU9 A0A1D1V988 A0A0V0V9N3 A0A2T7PQ59 F1KUD7 A0A0J7KPV1 A0A1Y3E554 A0A0V1G180 A0A0V1DZK2 A0A0V1IER3 A0A0V0S1G7 A0A0V0ZRI6 A0A183UHW9 A0A0N4UAR5 A0A267GZ12 A0A0V1CIL3 A0A0B2VYF2 A0A0V1CHV4 A0A0V1KZ58 A0A0V1L001 A0A0V0VA23 A0A0V1NTJ2
A0A0L7LAU3 A0A151IT52 A0A1Y1MW86 D6X1L3 A0A151IHA4 A0A087ZQ86 A0A1Y1MXK3 A0A195BX43 A0A3L8DI78 A0A2J7PTV3 A0A195F165 A0A026WT67 K7IRM5 A0A139WCH6 E2ACM2 A0A139WCL8 A0A067RB97 A0A1W4W8A5 A0A1W4WJK0 E2BNV4 A0A232EYW1 A0A2A3EIX6 A0A310STZ2 A0A139WCH5 A0A1B6EPD4 E0W0L9 A0A154P438 A0A1W4WIK8 A0A1W4WJU5 A0A0M9A2G0 A0A2J7PTV5 A0A151XG02 A0A0L7R4B5 A0A1Y1MZN3 A0A1Y1MW11 A0A1W4W8A3 A0A0C9RGY2 A0A0A9XTU7 A0A146M342 A0A1S3HIE6 A0A0P4VY24 V4A8J1 A0A3S5WGY9 A0A3S3NXN4 A0A210Q5K8 A0A0P4VT58 A0A023F0M5 A0A2P6KLZ8 A0A0P4W0C3 A0A2P8YRY4 A0A2C9K4L2 R7V8F5 T1IJA1 A0A0B7A9T4 A0A0L8FG06 A0A0K2V557 A0A0T6AWK9 T1ELW1 T1KXQ6 T1KXG2 A0A147BPZ7 A0A1Y1MW26 T1IBP8 L7MDZ2 A0A131YFM6 A0A1E1XHI1 F4WAU9 A0A132AD25 A0A0V1GZU4 A0A0V1MKU9 A0A1D1V988 A0A0V0V9N3 A0A2T7PQ59 F1KUD7 A0A0J7KPV1 A0A1Y3E554 A0A0V1G180 A0A0V1DZK2 A0A0V1IER3 A0A0V0S1G7 A0A0V0ZRI6 A0A183UHW9 A0A0N4UAR5 A0A267GZ12 A0A0V1CIL3 A0A0B2VYF2 A0A0V1CHV4 A0A0V1KZ58 A0A0V1L001 A0A0V0VA23 A0A0V1NTJ2
PDB
5JW7
E-value=1.26659e-09,
Score=153
Ontologies
GO
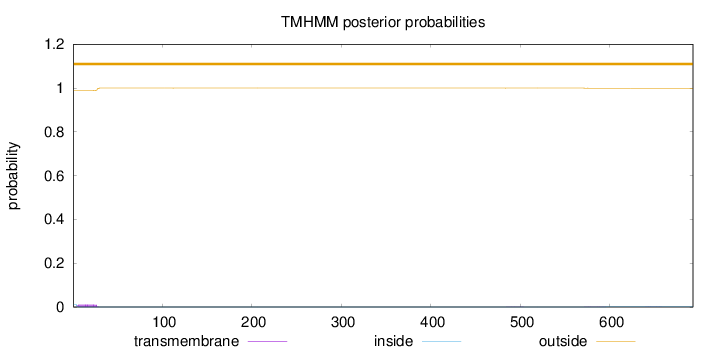
Topology
Length:
693
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.262850000000001
Exp number, first 60 AAs:
0.24718
Total prob of N-in:
0.01126
outside
1 - 693
Population Genetic Test Statistics
Pi
214.599922
Theta
152.789273
Tajima's D
1.049519
CLR
0.058497
CSRT
0.67891605419729
Interpretation
Uncertain
