Pre Gene Modal
BGIBMGA005453
Annotation
PREDICTED:_methyltransferase-like_protein_isoform_X2_[Papilio_machaon]
Full name
Methyltransferase-like protein
Location in the cell
Cytoplasmic Reliability : 2.223 Nuclear Reliability : 1.99
Sequence
CDS
ATGGTTGATAATTCTGATAAAAGACCTCAATTTGGGAATAGATATCTGGAAAATGCTGAAGAAGTGTTTAAACACAACTCGTGGGATAATGTGGAATGGGATGAAGACCAAGAGAAAAAAGCACAAGAGAAGGTTAGAAATAATTCACAAGTAACATTCACAGATGAACATTTAAAGTCCTTAGAAGAAGATGCTGATAAACACTGGGATGCATTCTATGATATACACCAAAACAGATTCTTCAAAGACCGGCACTGGCTTTTCACTGAATTTCCTGAGTTAGCACCTGACAATACATCTGCACCTGTACGAGTGTTTTCAGAAACTACAAACCATGCAAACAAACAAAACACAACTCTCAGTTCTACTAGTGATACAATTCGTAACATATTTGAAATAGGTTGTGGTGTTGGAAACACAATTTTCCCAATACTGCAATACAGTCAAGATCCAAAATTATTTGTATATGGCTGTGATTTCTCATCTAAAGCTATTGAAATAATGAAACAAAATGAGCTTTACAATACAGATAGATGTAAAGTATTTGTATTGGATGCAACAGAGGCTGACTGGAATGTACCTTTCAAGGAGAATTCAATAGATATTGTGGTGTTGATATTTGTATTGTCTGCTATTGACCCCCAAAAGATGCCTCAAGTAATCAAGAATATATTCAAATATCTGAAACCGGGTGGATTAGTTGTGTTCCGAGATTATGGCCTCTATGACCTAGCACAACTAAGGTTCAAAAAAGGAAGGTGTATATCCAACAACTTTTATGCACGAGGTGACAAAACCAGAGTATATTTTTTTACACAAGAAGAAATAAATAAACTCTTCACGGGTGAAGGATTCATAGAGGAGCAGAACCTTGTAGATAGAAGGCTACAAGTAAATAGAGGAAAGATGTTGACAATGTACAGAGTTTGGATACAAGCAAAATATAGAAAGCCGGTTTAA
Protein
MVDNSDKRPQFGNRYLENAEEVFKHNSWDNVEWDEDQEKKAQEKVRNNSQVTFTDEHLKSLEEDADKHWDAFYDIHQNRFFKDRHWLFTEFPELAPDNTSAPVRVFSETTNHANKQNTTLSSTSDTIRNIFEIGCGVGNTIFPILQYSQDPKLFVYGCDFSSKAIEIMKQNELYNTDRCKVFVLDATEADWNVPFKENSIDIVVLIFVLSAIDPQKMPQVIKNIFKYLKPGGLVVFRDYGLYDLAQLRFKKGRCISNNFYARGDKTRVYFFTQEEINKLFTGEGFIEEQNLVDRRLQVNRGKMLTMYRVWIQAKYRKPV
Summary
Subunit
Interacts with Psn.
Similarity
Belongs to the methyltransferase superfamily. METL family.
Keywords
Alternative splicing
Complete proteome
Methyltransferase
Reference proteome
Transferase
Feature
chain Methyltransferase-like protein
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9J7G1
A0A2H1WJD6
A0A194PTG0
A0A194QXY8
S4NZ65
A0A3S2NFA5
+ More
A0A212EU24 A0A0L7QM71 K7IQB9 A0A232EK54 A0A182P5I9 A0A182REG0 A0A023ER82 A0A182H599 E2C2L7 A0A1S4FFF8 A0A088APV2 Q7Q689 A0A182MHH2 A0A195CAP3 A0A182NIK8 A0A2A3EJZ5 A0A2M3ZBJ0 A0A182X736 Q0IF16 E2A5I8 A0A084W0Y1 A0A182G531 A0A1I8Q2P1 A0A0J7L193 A0A2M4BTH4 A0A151WTB4 A0A2M4BTI4 E9IDF5 A0A2M4BTL0 B0WPC5 A0A182J254 A0A195FQ86 A0A2M4BTF7 W5JAC7 A0A2M4BUE0 A0A0L7LQK0 A0A026WDQ1 A0A2M4ARE7 A0A0A1XIU1 A0A2M4ARE6 U5EU59 A0A034WVV5 A0A0K8WDU3 F4WJF6 A0A1I8M0G8 A0A1S4FFL5 A0A182YD02 W8BJJ9 A0A195BXB3 A0A2M3ZBT7 Q0IF15 E0VKH6 A0A182V3K0 A0A1Y1JU49 A0A1A9ZS30 A0A182K6S9 A0A182SFS9 A0A1A9YEH4 A0A0K8TQC0 A0A182QLF8 A0A1I8Q2M5 A0A182FGY6 A0A2J7QYH1 A0A0A1WFU1 A0A3B0J4G7 A0A034WUN1 A0A0K8VW48 A0A1A9WD23 A0A0P8XUR8 A0A2J7QYG6 Q29DZ8 B4N706 A0A3Q0ISY6 A0A1Y1JYY8 A0A2R7W0C6 A0A131XPI9 D6WUW9 A0A182WKM9 A0A1B0FP67 A0A0A9Y8V3 A0A0K8SLU7 B3NES1 Q86BS6-3 F2FB75 A0A1W4W890 A0A1B0B7X1 B4LBT3 B4NSZ4 A0A0M4ENY8 A0A224Z2S8 A0A336KEW9 A0A131XB80 A0A1W4XE47 B4HVW6
A0A212EU24 A0A0L7QM71 K7IQB9 A0A232EK54 A0A182P5I9 A0A182REG0 A0A023ER82 A0A182H599 E2C2L7 A0A1S4FFF8 A0A088APV2 Q7Q689 A0A182MHH2 A0A195CAP3 A0A182NIK8 A0A2A3EJZ5 A0A2M3ZBJ0 A0A182X736 Q0IF16 E2A5I8 A0A084W0Y1 A0A182G531 A0A1I8Q2P1 A0A0J7L193 A0A2M4BTH4 A0A151WTB4 A0A2M4BTI4 E9IDF5 A0A2M4BTL0 B0WPC5 A0A182J254 A0A195FQ86 A0A2M4BTF7 W5JAC7 A0A2M4BUE0 A0A0L7LQK0 A0A026WDQ1 A0A2M4ARE7 A0A0A1XIU1 A0A2M4ARE6 U5EU59 A0A034WVV5 A0A0K8WDU3 F4WJF6 A0A1I8M0G8 A0A1S4FFL5 A0A182YD02 W8BJJ9 A0A195BXB3 A0A2M3ZBT7 Q0IF15 E0VKH6 A0A182V3K0 A0A1Y1JU49 A0A1A9ZS30 A0A182K6S9 A0A182SFS9 A0A1A9YEH4 A0A0K8TQC0 A0A182QLF8 A0A1I8Q2M5 A0A182FGY6 A0A2J7QYH1 A0A0A1WFU1 A0A3B0J4G7 A0A034WUN1 A0A0K8VW48 A0A1A9WD23 A0A0P8XUR8 A0A2J7QYG6 Q29DZ8 B4N706 A0A3Q0ISY6 A0A1Y1JYY8 A0A2R7W0C6 A0A131XPI9 D6WUW9 A0A182WKM9 A0A1B0FP67 A0A0A9Y8V3 A0A0K8SLU7 B3NES1 Q86BS6-3 F2FB75 A0A1W4W890 A0A1B0B7X1 B4LBT3 B4NSZ4 A0A0M4ENY8 A0A224Z2S8 A0A336KEW9 A0A131XB80 A0A1W4XE47 B4HVW6
EC Number
2.1.1.-
Pubmed
19121390
26354079
23622113
22118469
20075255
28648823
+ More
24945155 26483478 20798317 12364791 14747013 17210077 17510324 24438588 21282665 20920257 23761445 26227816 24508170 30249741 25830018 25348373 21719571 25315136 25244985 24495485 20566863 28004739 26369729 17994087 15632085 18362917 19820115 25401762 26823975 10731132 12537572 12537569 11738826 22936249 28797301 28049606
24945155 26483478 20798317 12364791 14747013 17210077 17510324 24438588 21282665 20920257 23761445 26227816 24508170 30249741 25830018 25348373 21719571 25315136 25244985 24495485 20566863 28004739 26369729 17994087 15632085 18362917 19820115 25401762 26823975 10731132 12537572 12537569 11738826 22936249 28797301 28049606
EMBL
BABH01013785
BABH01013786
ODYU01009024
SOQ53127.1
KQ459599
KPI94420.1
+ More
KQ461154 KPJ08441.1 GAIX01011472 JAA81088.1 RSAL01000062 RVE49589.1 AGBW02012491 OWR44988.1 KQ414905 KOC59621.1 NNAY01003869 OXU18740.1 GAPW01002439 JAC11159.1 JXUM01026103 KQ560744 KXJ81035.1 GL452167 EFN77816.1 AAAB01008960 EAA11389.4 AXCM01007579 KQ978023 KYM97939.1 KZ288232 PBC31509.1 GGFM01005173 MBW25924.1 CH477420 EAT41257.1 GL436989 EFN71300.1 ATLV01019164 KE525263 KFB43875.1 JXUM01007162 KQ560234 KXJ83694.1 LBMM01001421 KMQ96353.1 GGFJ01007236 MBW56377.1 KQ982769 KYQ50875.1 GGFJ01007239 MBW56380.1 GL762454 EFZ21529.1 GGFJ01007238 MBW56379.1 DS232021 EDS32175.1 KQ981335 KYN42638.1 GGFJ01007235 MBW56376.1 ADMH02001844 ETN60926.1 GGFJ01007237 MBW56378.1 JTDY01000316 KOB77727.1 KK107260 QOIP01000001 EZA54182.1 RLU27220.1 GGFK01010030 MBW43351.1 GBXI01003366 JAD10926.1 GGFK01010046 MBW43367.1 GANO01002468 JAB57403.1 GAKP01000607 JAC58345.1 GDHF01002981 JAI49333.1 GL888182 EGI65734.1 GAMC01005115 JAC01441.1 KQ976401 KYM92591.1 GGFM01005184 MBW25935.1 EAT41258.1 DS235244 EEB13882.1 GEZM01102971 JAV51658.1 GDAI01001039 JAI16564.1 AXCN02000411 NEVH01009090 PNF33628.1 GBXI01016540 JAC97751.1 OUUW01000002 SPP76724.1 GAKP01000608 JAC58344.1 GDHF01009202 JAI43112.1 CH902618 KPU78474.1 PNF33630.1 CH379070 EAL30265.3 CH964168 EDW80145.2 GEZM01102972 JAV51657.1 KK854215 PTY13196.1 GEFM01006492 JAP69304.1 KQ971363 EFA09075.2 CCAG010018738 GBHO01016076 GBHO01016075 GDHC01002393 GDHC01001888 JAG27528.1 JAG27529.1 JAQ16236.1 JAQ16741.1 GBRD01012044 JAG53780.1 CH954178 EDV50194.1 AE014296 AY051875 BT126162 ADZ99414.1 JXJN01009733 CH940647 EDW68710.2 CH982512 CM002912 EDX15684.1 KMY96896.1 CP012525 ALC43742.1 GFPF01009727 MAA20873.1 UFQS01000340 UFQT01000340 SSX03001.1 SSX23368.1 GEFH01005163 JAP63418.1 CH480817 EDW50081.1
KQ461154 KPJ08441.1 GAIX01011472 JAA81088.1 RSAL01000062 RVE49589.1 AGBW02012491 OWR44988.1 KQ414905 KOC59621.1 NNAY01003869 OXU18740.1 GAPW01002439 JAC11159.1 JXUM01026103 KQ560744 KXJ81035.1 GL452167 EFN77816.1 AAAB01008960 EAA11389.4 AXCM01007579 KQ978023 KYM97939.1 KZ288232 PBC31509.1 GGFM01005173 MBW25924.1 CH477420 EAT41257.1 GL436989 EFN71300.1 ATLV01019164 KE525263 KFB43875.1 JXUM01007162 KQ560234 KXJ83694.1 LBMM01001421 KMQ96353.1 GGFJ01007236 MBW56377.1 KQ982769 KYQ50875.1 GGFJ01007239 MBW56380.1 GL762454 EFZ21529.1 GGFJ01007238 MBW56379.1 DS232021 EDS32175.1 KQ981335 KYN42638.1 GGFJ01007235 MBW56376.1 ADMH02001844 ETN60926.1 GGFJ01007237 MBW56378.1 JTDY01000316 KOB77727.1 KK107260 QOIP01000001 EZA54182.1 RLU27220.1 GGFK01010030 MBW43351.1 GBXI01003366 JAD10926.1 GGFK01010046 MBW43367.1 GANO01002468 JAB57403.1 GAKP01000607 JAC58345.1 GDHF01002981 JAI49333.1 GL888182 EGI65734.1 GAMC01005115 JAC01441.1 KQ976401 KYM92591.1 GGFM01005184 MBW25935.1 EAT41258.1 DS235244 EEB13882.1 GEZM01102971 JAV51658.1 GDAI01001039 JAI16564.1 AXCN02000411 NEVH01009090 PNF33628.1 GBXI01016540 JAC97751.1 OUUW01000002 SPP76724.1 GAKP01000608 JAC58344.1 GDHF01009202 JAI43112.1 CH902618 KPU78474.1 PNF33630.1 CH379070 EAL30265.3 CH964168 EDW80145.2 GEZM01102972 JAV51657.1 KK854215 PTY13196.1 GEFM01006492 JAP69304.1 KQ971363 EFA09075.2 CCAG010018738 GBHO01016076 GBHO01016075 GDHC01002393 GDHC01001888 JAG27528.1 JAG27529.1 JAQ16236.1 JAQ16741.1 GBRD01012044 JAG53780.1 CH954178 EDV50194.1 AE014296 AY051875 BT126162 ADZ99414.1 JXJN01009733 CH940647 EDW68710.2 CH982512 CM002912 EDX15684.1 KMY96896.1 CP012525 ALC43742.1 GFPF01009727 MAA20873.1 UFQS01000340 UFQT01000340 SSX03001.1 SSX23368.1 GEFH01005163 JAP63418.1 CH480817 EDW50081.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000283053
UP000007151
UP000053825
+ More
UP000002358 UP000215335 UP000075885 UP000075900 UP000069940 UP000249989 UP000008237 UP000005203 UP000007062 UP000075883 UP000078542 UP000075884 UP000242457 UP000076407 UP000008820 UP000000311 UP000030765 UP000095300 UP000036403 UP000075809 UP000002320 UP000075880 UP000078541 UP000000673 UP000037510 UP000053097 UP000279307 UP000007755 UP000095301 UP000076408 UP000078540 UP000009046 UP000075903 UP000092445 UP000075881 UP000075901 UP000092443 UP000075886 UP000069272 UP000235965 UP000268350 UP000091820 UP000007801 UP000001819 UP000007798 UP000079169 UP000007266 UP000075920 UP000092444 UP000008711 UP000000803 UP000192221 UP000092460 UP000008792 UP000000304 UP000092553 UP000192223 UP000001292
UP000002358 UP000215335 UP000075885 UP000075900 UP000069940 UP000249989 UP000008237 UP000005203 UP000007062 UP000075883 UP000078542 UP000075884 UP000242457 UP000076407 UP000008820 UP000000311 UP000030765 UP000095300 UP000036403 UP000075809 UP000002320 UP000075880 UP000078541 UP000000673 UP000037510 UP000053097 UP000279307 UP000007755 UP000095301 UP000076408 UP000078540 UP000009046 UP000075903 UP000092445 UP000075881 UP000075901 UP000092443 UP000075886 UP000069272 UP000235965 UP000268350 UP000091820 UP000007801 UP000001819 UP000007798 UP000079169 UP000007266 UP000075920 UP000092444 UP000008711 UP000000803 UP000192221 UP000092460 UP000008792 UP000000304 UP000092553 UP000192223 UP000001292
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9J7G1
A0A2H1WJD6
A0A194PTG0
A0A194QXY8
S4NZ65
A0A3S2NFA5
+ More
A0A212EU24 A0A0L7QM71 K7IQB9 A0A232EK54 A0A182P5I9 A0A182REG0 A0A023ER82 A0A182H599 E2C2L7 A0A1S4FFF8 A0A088APV2 Q7Q689 A0A182MHH2 A0A195CAP3 A0A182NIK8 A0A2A3EJZ5 A0A2M3ZBJ0 A0A182X736 Q0IF16 E2A5I8 A0A084W0Y1 A0A182G531 A0A1I8Q2P1 A0A0J7L193 A0A2M4BTH4 A0A151WTB4 A0A2M4BTI4 E9IDF5 A0A2M4BTL0 B0WPC5 A0A182J254 A0A195FQ86 A0A2M4BTF7 W5JAC7 A0A2M4BUE0 A0A0L7LQK0 A0A026WDQ1 A0A2M4ARE7 A0A0A1XIU1 A0A2M4ARE6 U5EU59 A0A034WVV5 A0A0K8WDU3 F4WJF6 A0A1I8M0G8 A0A1S4FFL5 A0A182YD02 W8BJJ9 A0A195BXB3 A0A2M3ZBT7 Q0IF15 E0VKH6 A0A182V3K0 A0A1Y1JU49 A0A1A9ZS30 A0A182K6S9 A0A182SFS9 A0A1A9YEH4 A0A0K8TQC0 A0A182QLF8 A0A1I8Q2M5 A0A182FGY6 A0A2J7QYH1 A0A0A1WFU1 A0A3B0J4G7 A0A034WUN1 A0A0K8VW48 A0A1A9WD23 A0A0P8XUR8 A0A2J7QYG6 Q29DZ8 B4N706 A0A3Q0ISY6 A0A1Y1JYY8 A0A2R7W0C6 A0A131XPI9 D6WUW9 A0A182WKM9 A0A1B0FP67 A0A0A9Y8V3 A0A0K8SLU7 B3NES1 Q86BS6-3 F2FB75 A0A1W4W890 A0A1B0B7X1 B4LBT3 B4NSZ4 A0A0M4ENY8 A0A224Z2S8 A0A336KEW9 A0A131XB80 A0A1W4XE47 B4HVW6
A0A212EU24 A0A0L7QM71 K7IQB9 A0A232EK54 A0A182P5I9 A0A182REG0 A0A023ER82 A0A182H599 E2C2L7 A0A1S4FFF8 A0A088APV2 Q7Q689 A0A182MHH2 A0A195CAP3 A0A182NIK8 A0A2A3EJZ5 A0A2M3ZBJ0 A0A182X736 Q0IF16 E2A5I8 A0A084W0Y1 A0A182G531 A0A1I8Q2P1 A0A0J7L193 A0A2M4BTH4 A0A151WTB4 A0A2M4BTI4 E9IDF5 A0A2M4BTL0 B0WPC5 A0A182J254 A0A195FQ86 A0A2M4BTF7 W5JAC7 A0A2M4BUE0 A0A0L7LQK0 A0A026WDQ1 A0A2M4ARE7 A0A0A1XIU1 A0A2M4ARE6 U5EU59 A0A034WVV5 A0A0K8WDU3 F4WJF6 A0A1I8M0G8 A0A1S4FFL5 A0A182YD02 W8BJJ9 A0A195BXB3 A0A2M3ZBT7 Q0IF15 E0VKH6 A0A182V3K0 A0A1Y1JU49 A0A1A9ZS30 A0A182K6S9 A0A182SFS9 A0A1A9YEH4 A0A0K8TQC0 A0A182QLF8 A0A1I8Q2M5 A0A182FGY6 A0A2J7QYH1 A0A0A1WFU1 A0A3B0J4G7 A0A034WUN1 A0A0K8VW48 A0A1A9WD23 A0A0P8XUR8 A0A2J7QYG6 Q29DZ8 B4N706 A0A3Q0ISY6 A0A1Y1JYY8 A0A2R7W0C6 A0A131XPI9 D6WUW9 A0A182WKM9 A0A1B0FP67 A0A0A9Y8V3 A0A0K8SLU7 B3NES1 Q86BS6-3 F2FB75 A0A1W4W890 A0A1B0B7X1 B4LBT3 B4NSZ4 A0A0M4ENY8 A0A224Z2S8 A0A336KEW9 A0A131XB80 A0A1W4XE47 B4HVW6
PDB
3MGG
E-value=0.000895444,
Score=99
Ontologies
PANTHER
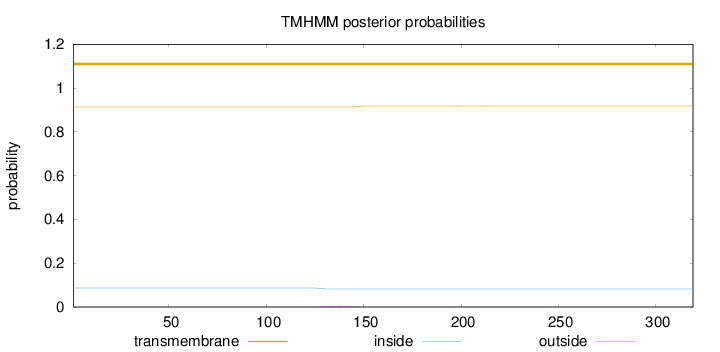
Topology
Length:
319
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10446
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08672
outside
1 - 319
Population Genetic Test Statistics
Pi
174.528685
Theta
144.195701
Tajima's D
0.796647
CLR
0.027573
CSRT
0.60021998900055
Interpretation
Uncertain
