Pre Gene Modal
BGIBMGA005452
Annotation
PREDICTED:_ribosome_biogenesis_protein_WDR12_homolog_[Amyelois_transitella]
Full name
Ribosome biogenesis protein WDR12 homolog
Location in the cell
Extracellular Reliability : 1.719
Sequence
CDS
ATGATCACTTACCGTGGCCACGACAAGGGAATTGAATGCCTGTCTGTGTCGGCTGATGGTAACCGTTTTGCAAGTGGGAGCTGGGATAACAATGTTTGTCTTTGGAGTGCAAGCTTAGCAGAAGAAGAAAACCATCCAGCTACGAAGAAAAGTAAACCAGAACACGGCACCACTAGAAATCCACTGACCACTTTAAAAGGACACAAGGAGGCTATCTCGGGTGTTCAATGGATGGATTACAACACAGTACTGTCCAGTGGTTGGGACCACTTATTGAAAATTTGGGACTGTGATTTGGGTGGTATTAAACAAGAAATTGCTGGTAATAAGGCCTTTTTTGATGTTGATTGGTCACCTCTGAACAACAGCATTATTACTGCATCAGCAGATAGACATGTCCGGTTATATGACCCTAGATCTACAGAGAGTATTGTGAAGACAACATTCACATCACACACAGGATGGGTCCAGTCAGTTCGCTGGTCTAAAACAAAAGACACTCTTTTCCTTTCTGCTGGATACGACAACCAGGTTAAACTTTGGGAAACAAGAAGTCCACGAACTCCACTTTATGACCTTAGCGGACACGAAGATAAAGTTCTCTGCTGCGATTGGTCGAATCCCTCCCTGTTAATCAGCGGCTCCAGCGACAACACCCTCAGGATATTCAAAGCTAAACACGCTGTCGGAAACGCGTGA
Protein
MITYRGHDKGIECLSVSADGNRFASGSWDNNVCLWSASLAEEENHPATKKSKPEHGTTRNPLTTLKGHKEAISGVQWMDYNTVLSSGWDHLLKIWDCDLGGIKQEIAGNKAFFDVDWSPLNNSIITASADRHVRLYDPRSTESIVKTTFTSHTGWVQSVRWSKTKDTLFLSAGYDNQVKLWETRSPRTPLYDLSGHEDKVLCCDWSNPSLLISGSSDNTLRIFKAKHAVGNA
Summary
Description
Required for maturation of ribosomal RNAs and formation of the large ribosomal subunit.
Similarity
Belongs to the WD repeat WDR12/YTM1 family.
Keywords
Complete proteome
Nucleus
Reference proteome
Repeat
Ribosome biogenesis
rRNA processing
WD repeat
Feature
chain Ribosome biogenesis protein WDR12 homolog
Uniprot
H9J7G0
A0A1E1WVF7
S4PAA1
A0A2H1VX79
A0A194PTG5
A0A194QSD8
+ More
A0A212EU55 A0A0M8ZQ61 A0A154PCV7 A0A0L7QLU0 A0A088APV0 K7IPU9 A0A2A3EIG1 A0A2J7PTS2 A0A2P8ZJU2 A0A067RI71 A0A336M4V3 A0A336MRQ0 A0A0J7KVS6 A0A195CCI8 A0A195BXA9 E2A5I5 A0A158NV27 F4WJF8 A0A182JK61 A0A1L8E5Q2 E9IDJ5 U5EMY7 A0A195FQE1 A0A1B6LNN0 A0A1L8E5J4 A0A232FKL2 A0A1B0D3H9 A0A182MM64 B0W517 A0A1J1IMX4 A0A182JPN2 A0A1S4J4Q8 A0A151WSV7 A0A1B6CDM3 A0A0P5GDL0 A0A084VNZ8 A0A1B6F462 E0W0L8 A0A1Q3G5G0 A0A0P5VES8 T1PJQ0 A0A0P4XXA8 A0A0P4Z3F0 A0A0P5KWT2 W8BY51 A0A182H105 A0A0P5GQ38 E2BUS4 A0A0A1WW16 A0A0P5TAV2 A0A182UCN1 A0A2C9GS25 Q7QJ33 A0A182KQQ1 A0A182YMN0 A0A182VC50 A0A0N8D4Y7 A0A182NKR6 Q17BB0 A0A0P5V3X0 A0A0N8CL69 A0A0P5W3D9 A0A1I8PIR2 A0A0N8CSN7 A0A0P6E3F7 E9GZX3 A0A0P5S6C9 A0A0P5YGU0 A0A0P5RWG4 A0A0P5H6D5 A0A0P5PG41 J9JKF4 A0A2S2PD99 A0A0P5UNK4 A0A0P6JV15 A0A226DZG9 A0A2H8TPF2 A0A0P5YTL3 A0A0P5NHJ9 A0A182QG95 A0A2M4AE07 A0A0P5ACV7 T1EA97 A0A0N8B493 A0A2M4ADT5 A0A2M4ADT1 A0A1D2N9P5 A0A182P250 A0A0K8VFN1 W5J4E0 A0A182XLL4 A0A1W4WZ08 A0A3B0JHC4 A0A0P5CKE9
A0A212EU55 A0A0M8ZQ61 A0A154PCV7 A0A0L7QLU0 A0A088APV0 K7IPU9 A0A2A3EIG1 A0A2J7PTS2 A0A2P8ZJU2 A0A067RI71 A0A336M4V3 A0A336MRQ0 A0A0J7KVS6 A0A195CCI8 A0A195BXA9 E2A5I5 A0A158NV27 F4WJF8 A0A182JK61 A0A1L8E5Q2 E9IDJ5 U5EMY7 A0A195FQE1 A0A1B6LNN0 A0A1L8E5J4 A0A232FKL2 A0A1B0D3H9 A0A182MM64 B0W517 A0A1J1IMX4 A0A182JPN2 A0A1S4J4Q8 A0A151WSV7 A0A1B6CDM3 A0A0P5GDL0 A0A084VNZ8 A0A1B6F462 E0W0L8 A0A1Q3G5G0 A0A0P5VES8 T1PJQ0 A0A0P4XXA8 A0A0P4Z3F0 A0A0P5KWT2 W8BY51 A0A182H105 A0A0P5GQ38 E2BUS4 A0A0A1WW16 A0A0P5TAV2 A0A182UCN1 A0A2C9GS25 Q7QJ33 A0A182KQQ1 A0A182YMN0 A0A182VC50 A0A0N8D4Y7 A0A182NKR6 Q17BB0 A0A0P5V3X0 A0A0N8CL69 A0A0P5W3D9 A0A1I8PIR2 A0A0N8CSN7 A0A0P6E3F7 E9GZX3 A0A0P5S6C9 A0A0P5YGU0 A0A0P5RWG4 A0A0P5H6D5 A0A0P5PG41 J9JKF4 A0A2S2PD99 A0A0P5UNK4 A0A0P6JV15 A0A226DZG9 A0A2H8TPF2 A0A0P5YTL3 A0A0P5NHJ9 A0A182QG95 A0A2M4AE07 A0A0P5ACV7 T1EA97 A0A0N8B493 A0A2M4ADT5 A0A2M4ADT1 A0A1D2N9P5 A0A182P250 A0A0K8VFN1 W5J4E0 A0A182XLL4 A0A1W4WZ08 A0A3B0JHC4 A0A0P5CKE9
Pubmed
EMBL
BABH01013775
GDQN01000044
JAT91010.1
GAIX01003344
JAA89216.1
ODYU01005000
+ More
SOQ45440.1 KQ459599 KPI94425.1 KQ461154 KPJ08438.1 AGBW02012491 OWR44991.1 KQ435922 KOX68667.1 KQ434874 KZC09682.1 KQ414905 KOC59618.1 KZ288232 PBC31507.1 NEVH01021221 PNF19730.1 PYGN01000035 PSN56766.1 KK852619 KDR20110.1 UFQS01000560 UFQT01000560 SSX04950.1 SSX25312.1 UFQT01001049 SSX28628.1 LBMM01002784 KMQ94369.1 KQ978023 KYM97941.1 KQ976401 KYM92593.1 GL436989 EFN71297.1 ADTU01026871 ADTU01026872 GL888182 EGI65736.1 GFDF01000242 JAV13842.1 GL762454 EFZ21373.1 GANO01000809 JAB59062.1 KQ981335 KYN42636.1 GEBQ01014699 GEBQ01006134 JAT25278.1 JAT33843.1 GFDF01000195 JAV13889.1 NNAY01000087 OXU31123.1 AJVK01010976 AXCM01002371 DS231840 CVRI01000055 CRL01098.1 KQ982769 KYQ50877.1 GEDC01025918 JAS11380.1 GDIQ01242454 JAK09271.1 ATLV01014965 KE524999 KFB39692.1 GECZ01024791 JAS44978.1 DS235861 EEB19174.1 GFDL01000010 JAV35035.1 GDIP01102803 JAM00912.1 KA648981 AFP63610.1 GDIP01235437 JAI87964.1 GDIP01218541 JAJ04861.1 GDIQ01186312 JAK65413.1 GAMC01002278 JAC04278.1 JXUM01102366 KQ564722 KXJ71840.1 GDIQ01239350 JAK12375.1 GL450746 EFN80547.1 GBXI01011659 JAD02633.1 GDIP01134803 JAL68911.1 APCN01000179 AAAB01008807 GDIP01068351 JAM35364.1 CH477324 GDIP01120896 JAL82818.1 GDIP01120895 JAL82819.1 GDIP01105600 GDIQ01061559 JAL98114.1 JAN33178.1 GDIP01102751 JAM00964.1 GDIP01136042 GDIQ01083103 JAL67672.1 JAN11634.1 GL732579 EFX74983.1 GDIQ01108898 JAL42828.1 GDIP01059610 JAM44105.1 GDIQ01095689 JAL56037.1 GDIQ01231656 JAK20069.1 GDIQ01139034 JAL12692.1 ABLF02004647 ABLF02004648 ABLF02059306 GGMR01014745 MBY27364.1 GDIP01114158 JAL89556.1 GDIQ01007746 JAN86991.1 LNIX01000009 OXA50091.1 GFXV01004252 MBW16057.1 GDIP01054821 JAM48894.1 GDIQ01152630 JAK99095.1 AXCN02000542 GGFK01005621 MBW38942.1 GDIP01204142 JAJ19260.1 GAMD01000622 JAB00969.1 GDIQ01214315 JAK37410.1 GGFK01005622 MBW38943.1 GGFK01005620 MBW38941.1 LJIJ01000132 ODN01974.1 GDHF01014637 JAI37677.1 ADMH02002093 ETN59327.1 OUUW01000006 SPP81747.1 GDIP01170343 JAJ53059.1
SOQ45440.1 KQ459599 KPI94425.1 KQ461154 KPJ08438.1 AGBW02012491 OWR44991.1 KQ435922 KOX68667.1 KQ434874 KZC09682.1 KQ414905 KOC59618.1 KZ288232 PBC31507.1 NEVH01021221 PNF19730.1 PYGN01000035 PSN56766.1 KK852619 KDR20110.1 UFQS01000560 UFQT01000560 SSX04950.1 SSX25312.1 UFQT01001049 SSX28628.1 LBMM01002784 KMQ94369.1 KQ978023 KYM97941.1 KQ976401 KYM92593.1 GL436989 EFN71297.1 ADTU01026871 ADTU01026872 GL888182 EGI65736.1 GFDF01000242 JAV13842.1 GL762454 EFZ21373.1 GANO01000809 JAB59062.1 KQ981335 KYN42636.1 GEBQ01014699 GEBQ01006134 JAT25278.1 JAT33843.1 GFDF01000195 JAV13889.1 NNAY01000087 OXU31123.1 AJVK01010976 AXCM01002371 DS231840 CVRI01000055 CRL01098.1 KQ982769 KYQ50877.1 GEDC01025918 JAS11380.1 GDIQ01242454 JAK09271.1 ATLV01014965 KE524999 KFB39692.1 GECZ01024791 JAS44978.1 DS235861 EEB19174.1 GFDL01000010 JAV35035.1 GDIP01102803 JAM00912.1 KA648981 AFP63610.1 GDIP01235437 JAI87964.1 GDIP01218541 JAJ04861.1 GDIQ01186312 JAK65413.1 GAMC01002278 JAC04278.1 JXUM01102366 KQ564722 KXJ71840.1 GDIQ01239350 JAK12375.1 GL450746 EFN80547.1 GBXI01011659 JAD02633.1 GDIP01134803 JAL68911.1 APCN01000179 AAAB01008807 GDIP01068351 JAM35364.1 CH477324 GDIP01120896 JAL82818.1 GDIP01120895 JAL82819.1 GDIP01105600 GDIQ01061559 JAL98114.1 JAN33178.1 GDIP01102751 JAM00964.1 GDIP01136042 GDIQ01083103 JAL67672.1 JAN11634.1 GL732579 EFX74983.1 GDIQ01108898 JAL42828.1 GDIP01059610 JAM44105.1 GDIQ01095689 JAL56037.1 GDIQ01231656 JAK20069.1 GDIQ01139034 JAL12692.1 ABLF02004647 ABLF02004648 ABLF02059306 GGMR01014745 MBY27364.1 GDIP01114158 JAL89556.1 GDIQ01007746 JAN86991.1 LNIX01000009 OXA50091.1 GFXV01004252 MBW16057.1 GDIP01054821 JAM48894.1 GDIQ01152630 JAK99095.1 AXCN02000542 GGFK01005621 MBW38942.1 GDIP01204142 JAJ19260.1 GAMD01000622 JAB00969.1 GDIQ01214315 JAK37410.1 GGFK01005622 MBW38943.1 GGFK01005620 MBW38941.1 LJIJ01000132 ODN01974.1 GDHF01014637 JAI37677.1 ADMH02002093 ETN59327.1 OUUW01000006 SPP81747.1 GDIP01170343 JAJ53059.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000053105
UP000076502
+ More
UP000053825 UP000005203 UP000002358 UP000242457 UP000235965 UP000245037 UP000027135 UP000036403 UP000078542 UP000078540 UP000000311 UP000005205 UP000007755 UP000075880 UP000078541 UP000215335 UP000092462 UP000075883 UP000002320 UP000183832 UP000075881 UP000075809 UP000030765 UP000009046 UP000095301 UP000069940 UP000249989 UP000008237 UP000075902 UP000075840 UP000007062 UP000075882 UP000076408 UP000075903 UP000075884 UP000008820 UP000095300 UP000000305 UP000007819 UP000198287 UP000075886 UP000094527 UP000075885 UP000000673 UP000076407 UP000192223 UP000268350
UP000053825 UP000005203 UP000002358 UP000242457 UP000235965 UP000245037 UP000027135 UP000036403 UP000078542 UP000078540 UP000000311 UP000005205 UP000007755 UP000075880 UP000078541 UP000215335 UP000092462 UP000075883 UP000002320 UP000183832 UP000075881 UP000075809 UP000030765 UP000009046 UP000095301 UP000069940 UP000249989 UP000008237 UP000075902 UP000075840 UP000007062 UP000075882 UP000076408 UP000075903 UP000075884 UP000008820 UP000095300 UP000000305 UP000007819 UP000198287 UP000075886 UP000094527 UP000075885 UP000000673 UP000076407 UP000192223 UP000268350
PRIDE
Pfam
Interpro
IPR017986
WD40_repeat_dom
+ More
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR028599 WDR12/Ytm1
IPR012972 NLE
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR000219 DH-domain
IPR035899 DBL_dom_sf
IPR001357 BRCT_dom
IPR026817 Ect2
IPR001331 GDS_CDC24_CS
IPR011993 PH-like_dom_sf
IPR036420 BRCT_dom_sf
IPR003960 ATPase_AAA_CS
IPR027417 P-loop_NTPase
IPR003959 ATPase_AAA_core
IPR005937 26S_Psome_P45-like
IPR032501 Prot_ATP_ID_OB
IPR003593 AAA+_ATPase
IPR041569 AAA_lid_3
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR028599 WDR12/Ytm1
IPR012972 NLE
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR000219 DH-domain
IPR035899 DBL_dom_sf
IPR001357 BRCT_dom
IPR026817 Ect2
IPR001331 GDS_CDC24_CS
IPR011993 PH-like_dom_sf
IPR036420 BRCT_dom_sf
IPR003960 ATPase_AAA_CS
IPR027417 P-loop_NTPase
IPR003959 ATPase_AAA_core
IPR005937 26S_Psome_P45-like
IPR032501 Prot_ATP_ID_OB
IPR003593 AAA+_ATPase
IPR041569 AAA_lid_3
ProteinModelPortal
H9J7G0
A0A1E1WVF7
S4PAA1
A0A2H1VX79
A0A194PTG5
A0A194QSD8
+ More
A0A212EU55 A0A0M8ZQ61 A0A154PCV7 A0A0L7QLU0 A0A088APV0 K7IPU9 A0A2A3EIG1 A0A2J7PTS2 A0A2P8ZJU2 A0A067RI71 A0A336M4V3 A0A336MRQ0 A0A0J7KVS6 A0A195CCI8 A0A195BXA9 E2A5I5 A0A158NV27 F4WJF8 A0A182JK61 A0A1L8E5Q2 E9IDJ5 U5EMY7 A0A195FQE1 A0A1B6LNN0 A0A1L8E5J4 A0A232FKL2 A0A1B0D3H9 A0A182MM64 B0W517 A0A1J1IMX4 A0A182JPN2 A0A1S4J4Q8 A0A151WSV7 A0A1B6CDM3 A0A0P5GDL0 A0A084VNZ8 A0A1B6F462 E0W0L8 A0A1Q3G5G0 A0A0P5VES8 T1PJQ0 A0A0P4XXA8 A0A0P4Z3F0 A0A0P5KWT2 W8BY51 A0A182H105 A0A0P5GQ38 E2BUS4 A0A0A1WW16 A0A0P5TAV2 A0A182UCN1 A0A2C9GS25 Q7QJ33 A0A182KQQ1 A0A182YMN0 A0A182VC50 A0A0N8D4Y7 A0A182NKR6 Q17BB0 A0A0P5V3X0 A0A0N8CL69 A0A0P5W3D9 A0A1I8PIR2 A0A0N8CSN7 A0A0P6E3F7 E9GZX3 A0A0P5S6C9 A0A0P5YGU0 A0A0P5RWG4 A0A0P5H6D5 A0A0P5PG41 J9JKF4 A0A2S2PD99 A0A0P5UNK4 A0A0P6JV15 A0A226DZG9 A0A2H8TPF2 A0A0P5YTL3 A0A0P5NHJ9 A0A182QG95 A0A2M4AE07 A0A0P5ACV7 T1EA97 A0A0N8B493 A0A2M4ADT5 A0A2M4ADT1 A0A1D2N9P5 A0A182P250 A0A0K8VFN1 W5J4E0 A0A182XLL4 A0A1W4WZ08 A0A3B0JHC4 A0A0P5CKE9
A0A212EU55 A0A0M8ZQ61 A0A154PCV7 A0A0L7QLU0 A0A088APV0 K7IPU9 A0A2A3EIG1 A0A2J7PTS2 A0A2P8ZJU2 A0A067RI71 A0A336M4V3 A0A336MRQ0 A0A0J7KVS6 A0A195CCI8 A0A195BXA9 E2A5I5 A0A158NV27 F4WJF8 A0A182JK61 A0A1L8E5Q2 E9IDJ5 U5EMY7 A0A195FQE1 A0A1B6LNN0 A0A1L8E5J4 A0A232FKL2 A0A1B0D3H9 A0A182MM64 B0W517 A0A1J1IMX4 A0A182JPN2 A0A1S4J4Q8 A0A151WSV7 A0A1B6CDM3 A0A0P5GDL0 A0A084VNZ8 A0A1B6F462 E0W0L8 A0A1Q3G5G0 A0A0P5VES8 T1PJQ0 A0A0P4XXA8 A0A0P4Z3F0 A0A0P5KWT2 W8BY51 A0A182H105 A0A0P5GQ38 E2BUS4 A0A0A1WW16 A0A0P5TAV2 A0A182UCN1 A0A2C9GS25 Q7QJ33 A0A182KQQ1 A0A182YMN0 A0A182VC50 A0A0N8D4Y7 A0A182NKR6 Q17BB0 A0A0P5V3X0 A0A0N8CL69 A0A0P5W3D9 A0A1I8PIR2 A0A0N8CSN7 A0A0P6E3F7 E9GZX3 A0A0P5S6C9 A0A0P5YGU0 A0A0P5RWG4 A0A0P5H6D5 A0A0P5PG41 J9JKF4 A0A2S2PD99 A0A0P5UNK4 A0A0P6JV15 A0A226DZG9 A0A2H8TPF2 A0A0P5YTL3 A0A0P5NHJ9 A0A182QG95 A0A2M4AE07 A0A0P5ACV7 T1EA97 A0A0N8B493 A0A2M4ADT5 A0A2M4ADT1 A0A1D2N9P5 A0A182P250 A0A0K8VFN1 W5J4E0 A0A182XLL4 A0A1W4WZ08 A0A3B0JHC4 A0A0P5CKE9
PDB
6N31
E-value=1.61936e-50,
Score=501
Ontologies
KEGG
GO
GO:0000466
GO:0005654
GO:0000463
GO:0043021
GO:0005730
GO:0030687
GO:0042254
GO:0006364
GO:0005089
GO:0035556
GO:2000431
GO:0005096
GO:0035023
GO:0005737
GO:0070545
GO:0036402
GO:0030163
GO:0005524
GO:0005515
GO:0046872
GO:0006281
GO:0016887
GO:0030870
GO:0006310
GO:0006418
GO:0009058
GO:0016742
GO:0003824
PANTHER
Topology
Subcellular location
Nucleus
Nucleolus
Nucleoplasm
Nucleolus
Nucleoplasm
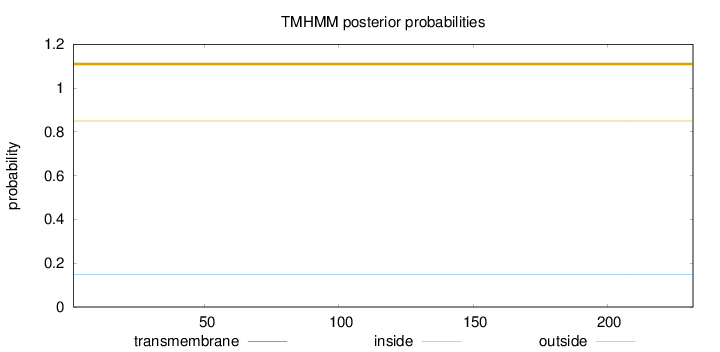
Length:
232
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00013
Exp number, first 60 AAs:
0
Total prob of N-in:
0.15003
outside
1 - 232
Population Genetic Test Statistics
Pi
229.030651
Theta
199.29392
Tajima's D
0.466309
CLR
0.033259
CSRT
0.498525073746313
Interpretation
Uncertain
