Pre Gene Modal
BGIBMGA005452
Annotation
PREDICTED:_ribosome_biogenesis_protein_WDR12_homolog_[Amyelois_transitella]
Full name
Ribosome biogenesis protein WDR12 homolog
Location in the cell
Extracellular Reliability : 2.076
Sequence
CDS
ATGTCTGGCGATGCTTTAGAAGCTCAATTGCAAGTTAGATTTATAACTAAACAAGAACAATATGCAGTTCCGGATAGTCCTTACGCTATCCAGTGTAACGTACACCCTGCGGACCTTAATACCTTAATCAACGCAATTTTGAAGGAGACCTCACCATCGTTCGAGAAGGCGGTAATATTTGATTTCTTAGTATGTGGAGAGTTACTTTGTGCCTCCCTCGCAGAGCACTTACAAGAAAAAGGGGTATCGACAGAGGACACGCTTGAGGTCGAATACTTGGAAAGGTTCCCTGCACCAACTCCACAGGACTGTTTAATGCACGATGATTGGGTGTCAGCTGTTCAGGCACACAGTAGTTGGATTCTATCAGGAAGTTATGACAATAGCTTACATATATGGAGCACAAAAGGACAACACAAACTAGCCATACCAGGTCATACATCACCAGTAAAAGCAGTATCATGGGTGTCCTTCCAGGGTGATCAAGCTACATTTGTTAGATATAGCATAGCAGTTATGATCTGA
Protein
MSGDALEAQLQVRFITKQEQYAVPDSPYAIQCNVHPADLNTLINAILKETSPSFEKAVIFDFLVCGELLCASLAEHLQEKGVSTEDTLEVEYLERFPAPTPQDCLMHDDWVSAVQAHSSWILSGSYDNSLHIWSTKGQHKLAIPGHTSPVKAVSWVSFQGDQATFVRYSIAVMI
Summary
Description
Required for maturation of ribosomal RNAs and formation of the large ribosomal subunit.
Similarity
Belongs to the WD repeat WDR12/YTM1 family.
Belongs to the ubiquitin-conjugating enzyme family.
Belongs to the ubiquitin-conjugating enzyme family.
Keywords
Complete proteome
Nucleus
Reference proteome
Repeat
Ribosome biogenesis
rRNA processing
WD repeat
Feature
chain Ribosome biogenesis protein WDR12 homolog
Uniprot
H9J7G0
A0A2A4IU73
A0A2H1VX79
A0A1E1WVF7
I4DPL0
A0A194PTG5
+ More
S4PAA1 A0A194QSD8 A0A212EU55 A0A1B6J785 A0A1B6LGL4 A0A1B6F462 A0A182MM64 A0A1B6CDM3 A0A0J7KVS6 A0A182YMN0 K7IPU9 A0A182P250 A0A182UCN1 A0A2C9GS25 Q7QJ33 A0A182VC50 A0A1L8E5J4 A0A023ERD6 A0A0M8ZQ61 A0A182KQQ1 E9IDJ5 A0A154PCV7 Q17BB0 A0A088APV0 A0A1Q3G5G0 Q29KQ0 A0A182NKR6 B4GT01 F4WJF8 A0A3B0JHC4 B4KKN1 W5J4E0 A0A195BXA9 W8BY51 A0A182QG95 A0A182XLL4 A0A1B0D3H9 A0A195FQE1 A0A151WSV7 A0A158NV27 A0A2M4ADT5 A0A2P8ZJU2 A0A1I8PIR2 A0A2A3EIG1 B0W517 A0A2M4ADT1 A0A2M4AE07 A0A182JK61 B4HWV6 B4MU54 A0A182JPN2 A0A232FKL2 U5EMY7 A0A195CCI8 A0A2M4BNU7 A0A1W4VSY1 A0A0J9TKT5 B4Q9T6 A0A182F4J1 A0A1A9WZX7 A0A2J7PTS2 Q9VKQ3 M9PB96 A0A0M4EPV5 B4LS78 A0A1A9UMI4 A0A170YP47 A0A1A9XZJ1 A0A1B0B919 B3MJV8 A0A1A9Z7S1 D3TPJ6 A0A1B0G5Y7 T1PJQ0 A0A182R3H8 B4P116 B3N534 A0A0A1WW16 A0A336M4V3 A0A336MRQ0 E0W0L8 B4JPT9 A0A0L0BZH2 A0A0K8VFN1 E9GZX3 A0A182WJQ3 A0A0T6B3S7 T1I4N8 T1EA97 A0A0V0G7F4 A0A034WWZ6 A0A2S2PD99 A0A069DUD1 A0A0L7QLU0
S4PAA1 A0A194QSD8 A0A212EU55 A0A1B6J785 A0A1B6LGL4 A0A1B6F462 A0A182MM64 A0A1B6CDM3 A0A0J7KVS6 A0A182YMN0 K7IPU9 A0A182P250 A0A182UCN1 A0A2C9GS25 Q7QJ33 A0A182VC50 A0A1L8E5J4 A0A023ERD6 A0A0M8ZQ61 A0A182KQQ1 E9IDJ5 A0A154PCV7 Q17BB0 A0A088APV0 A0A1Q3G5G0 Q29KQ0 A0A182NKR6 B4GT01 F4WJF8 A0A3B0JHC4 B4KKN1 W5J4E0 A0A195BXA9 W8BY51 A0A182QG95 A0A182XLL4 A0A1B0D3H9 A0A195FQE1 A0A151WSV7 A0A158NV27 A0A2M4ADT5 A0A2P8ZJU2 A0A1I8PIR2 A0A2A3EIG1 B0W517 A0A2M4ADT1 A0A2M4AE07 A0A182JK61 B4HWV6 B4MU54 A0A182JPN2 A0A232FKL2 U5EMY7 A0A195CCI8 A0A2M4BNU7 A0A1W4VSY1 A0A0J9TKT5 B4Q9T6 A0A182F4J1 A0A1A9WZX7 A0A2J7PTS2 Q9VKQ3 M9PB96 A0A0M4EPV5 B4LS78 A0A1A9UMI4 A0A170YP47 A0A1A9XZJ1 A0A1B0B919 B3MJV8 A0A1A9Z7S1 D3TPJ6 A0A1B0G5Y7 T1PJQ0 A0A182R3H8 B4P116 B3N534 A0A0A1WW16 A0A336M4V3 A0A336MRQ0 E0W0L8 B4JPT9 A0A0L0BZH2 A0A0K8VFN1 E9GZX3 A0A182WJQ3 A0A0T6B3S7 T1I4N8 T1EA97 A0A0V0G7F4 A0A034WWZ6 A0A2S2PD99 A0A069DUD1 A0A0L7QLU0
Pubmed
19121390
22651552
26354079
23622113
22118469
25244985
+ More
20075255 12364791 24945155 20966253 21282665 17510324 15632085 17994087 21719571 20920257 23761445 24495485 21347285 29403074 28648823 22936249 10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867 20353571 25315136 14525923 25830018 20566863 26108605 21292972 25348373 26334808
20075255 12364791 24945155 20966253 21282665 17510324 15632085 17994087 21719571 20920257 23761445 24495485 21347285 29403074 28648823 22936249 10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867 20353571 25315136 14525923 25830018 20566863 26108605 21292972 25348373 26334808
EMBL
BABH01013775
NWSH01007981
PCG62732.1
ODYU01005000
SOQ45440.1
GDQN01000044
+ More
JAT91010.1 AK403658 BAM19850.1 KQ459599 KPI94425.1 GAIX01003344 JAA89216.1 KQ461154 KPJ08438.1 AGBW02012491 OWR44991.1 GECU01012682 JAS95024.1 GEBQ01017142 JAT22835.1 GECZ01024791 JAS44978.1 AXCM01002371 GEDC01025918 JAS11380.1 LBMM01002784 KMQ94369.1 APCN01000179 AAAB01008807 GFDF01000195 JAV13889.1 GAPW01002369 JAC11229.1 KQ435922 KOX68667.1 GL762454 EFZ21373.1 KQ434874 KZC09682.1 CH477324 GFDL01000010 JAV35035.1 CH379061 CH479189 GL888182 EGI65736.1 OUUW01000006 SPP81747.1 CH933807 ADMH02002093 ETN59327.1 KQ976401 KYM92593.1 GAMC01002278 JAC04278.1 AXCN02000542 AJVK01010976 KQ981335 KYN42636.1 KQ982769 KYQ50877.1 ADTU01026871 ADTU01026872 GGFK01005622 MBW38943.1 PYGN01000035 PSN56766.1 KZ288232 PBC31507.1 DS231840 GGFK01005620 MBW38941.1 GGFK01005621 MBW38942.1 CH480818 CH963852 NNAY01000087 OXU31123.1 GANO01000809 JAB59062.1 KQ978023 KYM97941.1 GGFJ01005502 MBW54643.1 CM002910 KMY89620.1 CM000361 NEVH01021221 PNF19730.1 AE014134 AY118871 AGB92898.1 CP012523 ALC38682.1 CH940649 GEMB01003104 JAS00105.1 JXJN01010184 JXJN01010185 CH902620 EZ423348 ADD19624.1 CCAG010022414 KA648981 AFP63610.1 CM000157 AY231921 CH954177 GBXI01011659 JAD02633.1 UFQS01000560 UFQT01000560 SSX04950.1 SSX25312.1 UFQT01001049 SSX28628.1 DS235861 EEB19174.1 CH916372 JRES01001119 KNC25413.1 GDHF01014637 JAI37677.1 GL732579 EFX74983.1 LJIG01015976 KRT81986.1 ACPB03009352 GAMD01000622 JAB00969.1 GECL01002206 JAP03918.1 GAKP01000242 JAC58710.1 GGMR01014745 MBY27364.1 GBGD01001508 JAC87381.1 KQ414905 KOC59618.1
JAT91010.1 AK403658 BAM19850.1 KQ459599 KPI94425.1 GAIX01003344 JAA89216.1 KQ461154 KPJ08438.1 AGBW02012491 OWR44991.1 GECU01012682 JAS95024.1 GEBQ01017142 JAT22835.1 GECZ01024791 JAS44978.1 AXCM01002371 GEDC01025918 JAS11380.1 LBMM01002784 KMQ94369.1 APCN01000179 AAAB01008807 GFDF01000195 JAV13889.1 GAPW01002369 JAC11229.1 KQ435922 KOX68667.1 GL762454 EFZ21373.1 KQ434874 KZC09682.1 CH477324 GFDL01000010 JAV35035.1 CH379061 CH479189 GL888182 EGI65736.1 OUUW01000006 SPP81747.1 CH933807 ADMH02002093 ETN59327.1 KQ976401 KYM92593.1 GAMC01002278 JAC04278.1 AXCN02000542 AJVK01010976 KQ981335 KYN42636.1 KQ982769 KYQ50877.1 ADTU01026871 ADTU01026872 GGFK01005622 MBW38943.1 PYGN01000035 PSN56766.1 KZ288232 PBC31507.1 DS231840 GGFK01005620 MBW38941.1 GGFK01005621 MBW38942.1 CH480818 CH963852 NNAY01000087 OXU31123.1 GANO01000809 JAB59062.1 KQ978023 KYM97941.1 GGFJ01005502 MBW54643.1 CM002910 KMY89620.1 CM000361 NEVH01021221 PNF19730.1 AE014134 AY118871 AGB92898.1 CP012523 ALC38682.1 CH940649 GEMB01003104 JAS00105.1 JXJN01010184 JXJN01010185 CH902620 EZ423348 ADD19624.1 CCAG010022414 KA648981 AFP63610.1 CM000157 AY231921 CH954177 GBXI01011659 JAD02633.1 UFQS01000560 UFQT01000560 SSX04950.1 SSX25312.1 UFQT01001049 SSX28628.1 DS235861 EEB19174.1 CH916372 JRES01001119 KNC25413.1 GDHF01014637 JAI37677.1 GL732579 EFX74983.1 LJIG01015976 KRT81986.1 ACPB03009352 GAMD01000622 JAB00969.1 GECL01002206 JAP03918.1 GAKP01000242 JAC58710.1 GGMR01014745 MBY27364.1 GBGD01001508 JAC87381.1 KQ414905 KOC59618.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000075883
+ More
UP000036403 UP000076408 UP000002358 UP000075885 UP000075902 UP000075840 UP000007062 UP000075903 UP000053105 UP000075882 UP000076502 UP000008820 UP000005203 UP000001819 UP000075884 UP000008744 UP000007755 UP000268350 UP000009192 UP000000673 UP000078540 UP000075886 UP000076407 UP000092462 UP000078541 UP000075809 UP000005205 UP000245037 UP000095300 UP000242457 UP000002320 UP000075880 UP000001292 UP000007798 UP000075881 UP000215335 UP000078542 UP000192221 UP000000304 UP000069272 UP000091820 UP000235965 UP000000803 UP000092553 UP000008792 UP000078200 UP000092443 UP000092460 UP000007801 UP000092445 UP000092444 UP000095301 UP000075900 UP000002282 UP000008711 UP000009046 UP000001070 UP000037069 UP000000305 UP000075920 UP000015103 UP000053825
UP000036403 UP000076408 UP000002358 UP000075885 UP000075902 UP000075840 UP000007062 UP000075903 UP000053105 UP000075882 UP000076502 UP000008820 UP000005203 UP000001819 UP000075884 UP000008744 UP000007755 UP000268350 UP000009192 UP000000673 UP000078540 UP000075886 UP000076407 UP000092462 UP000078541 UP000075809 UP000005205 UP000245037 UP000095300 UP000242457 UP000002320 UP000075880 UP000001292 UP000007798 UP000075881 UP000215335 UP000078542 UP000192221 UP000000304 UP000069272 UP000091820 UP000235965 UP000000803 UP000092553 UP000008792 UP000078200 UP000092443 UP000092460 UP000007801 UP000092445 UP000092444 UP000095301 UP000075900 UP000002282 UP000008711 UP000009046 UP000001070 UP000037069 UP000000305 UP000075920 UP000015103 UP000053825
Pfam
Interpro
IPR017986
WD40_repeat_dom
+ More
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR028599 WDR12/Ytm1
IPR012972 NLE
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR003960 ATPase_AAA_CS
IPR027417 P-loop_NTPase
IPR003959 ATPase_AAA_core
IPR005937 26S_Psome_P45-like
IPR032501 Prot_ATP_ID_OB
IPR003593 AAA+_ATPase
IPR041569 AAA_lid_3
IPR000219 DH-domain
IPR035899 DBL_dom_sf
IPR001357 BRCT_dom
IPR026817 Ect2
IPR001331 GDS_CDC24_CS
IPR011993 PH-like_dom_sf
IPR036420 BRCT_dom_sf
IPR016135 UBQ-conjugating_enzyme/RWD
IPR000608 UBQ-conjugat_E2
IPR023313 UBQ-conjugating_AS
IPR027012 Enkurin_dom
IPR016706 Cleav_polyA_spec_factor_su5
IPR000086 NUDIX_hydrolase_dom
IPR001163 LSM_dom_euk/arc
IPR010920 LSM_dom_sf
IPR034102 Sm_D1
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR028599 WDR12/Ytm1
IPR012972 NLE
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR003960 ATPase_AAA_CS
IPR027417 P-loop_NTPase
IPR003959 ATPase_AAA_core
IPR005937 26S_Psome_P45-like
IPR032501 Prot_ATP_ID_OB
IPR003593 AAA+_ATPase
IPR041569 AAA_lid_3
IPR000219 DH-domain
IPR035899 DBL_dom_sf
IPR001357 BRCT_dom
IPR026817 Ect2
IPR001331 GDS_CDC24_CS
IPR011993 PH-like_dom_sf
IPR036420 BRCT_dom_sf
IPR016135 UBQ-conjugating_enzyme/RWD
IPR000608 UBQ-conjugat_E2
IPR023313 UBQ-conjugating_AS
IPR027012 Enkurin_dom
IPR016706 Cleav_polyA_spec_factor_su5
IPR000086 NUDIX_hydrolase_dom
IPR001163 LSM_dom_euk/arc
IPR010920 LSM_dom_sf
IPR034102 Sm_D1
SUPFAM
ProteinModelPortal
H9J7G0
A0A2A4IU73
A0A2H1VX79
A0A1E1WVF7
I4DPL0
A0A194PTG5
+ More
S4PAA1 A0A194QSD8 A0A212EU55 A0A1B6J785 A0A1B6LGL4 A0A1B6F462 A0A182MM64 A0A1B6CDM3 A0A0J7KVS6 A0A182YMN0 K7IPU9 A0A182P250 A0A182UCN1 A0A2C9GS25 Q7QJ33 A0A182VC50 A0A1L8E5J4 A0A023ERD6 A0A0M8ZQ61 A0A182KQQ1 E9IDJ5 A0A154PCV7 Q17BB0 A0A088APV0 A0A1Q3G5G0 Q29KQ0 A0A182NKR6 B4GT01 F4WJF8 A0A3B0JHC4 B4KKN1 W5J4E0 A0A195BXA9 W8BY51 A0A182QG95 A0A182XLL4 A0A1B0D3H9 A0A195FQE1 A0A151WSV7 A0A158NV27 A0A2M4ADT5 A0A2P8ZJU2 A0A1I8PIR2 A0A2A3EIG1 B0W517 A0A2M4ADT1 A0A2M4AE07 A0A182JK61 B4HWV6 B4MU54 A0A182JPN2 A0A232FKL2 U5EMY7 A0A195CCI8 A0A2M4BNU7 A0A1W4VSY1 A0A0J9TKT5 B4Q9T6 A0A182F4J1 A0A1A9WZX7 A0A2J7PTS2 Q9VKQ3 M9PB96 A0A0M4EPV5 B4LS78 A0A1A9UMI4 A0A170YP47 A0A1A9XZJ1 A0A1B0B919 B3MJV8 A0A1A9Z7S1 D3TPJ6 A0A1B0G5Y7 T1PJQ0 A0A182R3H8 B4P116 B3N534 A0A0A1WW16 A0A336M4V3 A0A336MRQ0 E0W0L8 B4JPT9 A0A0L0BZH2 A0A0K8VFN1 E9GZX3 A0A182WJQ3 A0A0T6B3S7 T1I4N8 T1EA97 A0A0V0G7F4 A0A034WWZ6 A0A2S2PD99 A0A069DUD1 A0A0L7QLU0
S4PAA1 A0A194QSD8 A0A212EU55 A0A1B6J785 A0A1B6LGL4 A0A1B6F462 A0A182MM64 A0A1B6CDM3 A0A0J7KVS6 A0A182YMN0 K7IPU9 A0A182P250 A0A182UCN1 A0A2C9GS25 Q7QJ33 A0A182VC50 A0A1L8E5J4 A0A023ERD6 A0A0M8ZQ61 A0A182KQQ1 E9IDJ5 A0A154PCV7 Q17BB0 A0A088APV0 A0A1Q3G5G0 Q29KQ0 A0A182NKR6 B4GT01 F4WJF8 A0A3B0JHC4 B4KKN1 W5J4E0 A0A195BXA9 W8BY51 A0A182QG95 A0A182XLL4 A0A1B0D3H9 A0A195FQE1 A0A151WSV7 A0A158NV27 A0A2M4ADT5 A0A2P8ZJU2 A0A1I8PIR2 A0A2A3EIG1 B0W517 A0A2M4ADT1 A0A2M4AE07 A0A182JK61 B4HWV6 B4MU54 A0A182JPN2 A0A232FKL2 U5EMY7 A0A195CCI8 A0A2M4BNU7 A0A1W4VSY1 A0A0J9TKT5 B4Q9T6 A0A182F4J1 A0A1A9WZX7 A0A2J7PTS2 Q9VKQ3 M9PB96 A0A0M4EPV5 B4LS78 A0A1A9UMI4 A0A170YP47 A0A1A9XZJ1 A0A1B0B919 B3MJV8 A0A1A9Z7S1 D3TPJ6 A0A1B0G5Y7 T1PJQ0 A0A182R3H8 B4P116 B3N534 A0A0A1WW16 A0A336M4V3 A0A336MRQ0 E0W0L8 B4JPT9 A0A0L0BZH2 A0A0K8VFN1 E9GZX3 A0A182WJQ3 A0A0T6B3S7 T1I4N8 T1EA97 A0A0V0G7F4 A0A034WWZ6 A0A2S2PD99 A0A069DUD1 A0A0L7QLU0
PDB
6EM5
E-value=4.95133e-19,
Score=227
Ontologies
KEGG
GO
GO:0000466
GO:0005654
GO:0000463
GO:0043021
GO:0005730
GO:0030687
GO:0042254
GO:0070545
GO:0006364
GO:0036402
GO:0030163
GO:0005737
GO:0005524
GO:0005089
GO:0035556
GO:2000431
GO:0005096
GO:0035023
GO:0016740
GO:0005886
GO:0005829
GO:0006378
GO:0005849
GO:0003729
GO:0000387
GO:0005515
GO:0046872
GO:0006281
GO:0016887
GO:0030870
GO:0006310
GO:0006418
GO:0009058
GO:0016742
GO:0003824
PANTHER
Topology
Subcellular location
Nucleus
Nucleolus
Nucleoplasm
Nucleolus
Nucleoplasm
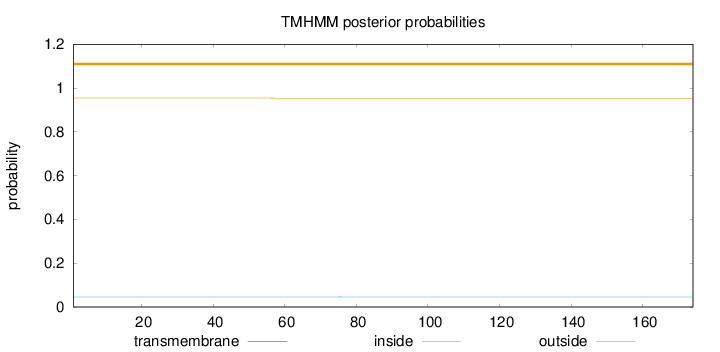
Length:
174
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0117
Exp number, first 60 AAs:
0.00225
Total prob of N-in:
0.04566
outside
1 - 174
Population Genetic Test Statistics
Pi
133.379087
Theta
153.763935
Tajima's D
-0.414934
CLR
0.427638
CSRT
0.266736663166842
Interpretation
Uncertain
