Gene
KWMTBOMO04510 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005358
Annotation
PREDICTED:_uncharacterized_protein_LOC101745349_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.98 Nuclear Reliability : 1.94
Sequence
CDS
ATGATATCGATGGCGCCGCCATATATGACACCTCCGGTAAGTCTCCTCTGTACAACATTTTTACAAGAACTAGCTGGCCAAGAAGAGCCCAAAGATGACGAAGTCACAGACCTCGAGGAGAAAATGAACGTAGATCAGTCTAGCGATGAAGAAGATGATGTAATAAAATTAAACGGCCCCTATGCACCAAGAGCAACCGAACTCTGGACACCGAATTACGAAGCCGTCAAAGCAAAGAAACTTAACAGAATACTTGAAGATCCGTTCATAGATTTGCACGCGACGAGTTCTTGGTGCATTGAAGAGTTTCCTAATAAAGTCATCGGATACCTTGGAGATCATCTTAAACTTGTTATTCAAGTCGAATCAAAAGGGATCCGGACTAATTTAAACTTGTTTATAAAATGCATGCCGAGGCTGGACAAGTGGAAAGCTCAGTATTTAAAAGAACTGAACTTCTTCAAAAAAGAGTATGTTATGCTCAGTAAATTATTCAAAGATTTTCGGAGCATTGAAGCTTCCCGTAAGTGGCGCCCTAAGCTACTCTTCATAAAAGAGGATATATTCGTTTTTGAAAATATCACAACAATTGGATATTCCATGCCAGACCACAAGGAGACCATGCCATTAGAAATGTTAAGAGCCGCTGTTAAAAGTTTAGCTAGATTCCACGCTGAATCATATATTTACGAGGAAAATAAATCTAAAGAACTCGGAAGACCCTACAGAATTTGGGAAGACTATAGCGAATACCTCCAAGAACCTGAACAAGGAATGGCCTGGCGGAATACTGGCAGAAACGGTGTTATTGATTTCTTAAAAGTGTTTTCAAGGTTTAAAGCCGAATCAAATTTTTCTCGACATTTAGATTTCATCATACCAATGCTATATGATGGAGCATTAGCTCTAATGAAACCCAGCTCTGAATACAGGAACGTAGTGGCACATCGTGATCTGTGGACGAACAATATACTGTTAAAAAAATGTGAAGATTCTTATCATGCTGTTTTAGTTGACTTTCAAACAGTCTTGTATTGTTCTCCGATGCTGGACTTGTCATCTCTCATATTCTTTAACACGACCAGAAGTGACAGATACAATTATACAGACGAACTCATCGATCTTTATTATGACACGATATCGGAAGAATTACTCATAAAAGATATTCAAATCAATGAAATCATGGACAAAGATACCTTGATAAAGGCGTACAATGATAGCATTATGTTTGGAATTACCCAAGCTGCGTTAATAGTTCCAATAGTATCGATGACCGAAGAGAAGAGAAAACAAATATTCTATGACCCTGAAAATTGTAAAAAAGCGAATGTCGTTTCTCGTGGTGAATACTTTATTGAAATCGCCAAAGAAGATGCATCTTATCGAAAAAGGGTACTAGAATTATTCGATGAAATTATTGAAAGATACATATATCCACATGAAAGCAGCCAACATAGACCCGATAAACATTAA
Protein
MISMAPPYMTPPVSLLCTTFLQELAGQEEPKDDEVTDLEEKMNVDQSSDEEDDVIKLNGPYAPRATELWTPNYEAVKAKKLNRILEDPFIDLHATSSWCIEEFPNKVIGYLGDHLKLVIQVESKGIRTNLNLFIKCMPRLDKWKAQYLKELNFFKKEYVMLSKLFKDFRSIEASRKWRPKLLFIKEDIFVFENITTIGYSMPDHKETMPLEMLRAAVKSLARFHAESYIYEENKSKELGRPYRIWEDYSEYLQEPEQGMAWRNTGRNGVIDFLKVFSRFKAESNFSRHLDFIIPMLYDGALALMKPSSEYRNVVAHRDLWTNNILLKKCEDSYHAVLVDFQTVLYCSPMLDLSSLIFFNTTRSDRYNYTDELIDLYYDTISEELLIKDIQINEIMDKDTLIKAYNDSIMFGITQAALIVPIVSMTEEKRKQIFYDPENCKKANVVSRGEYFIEIAKEDASYRKRVLELFDEIIERYIYPHESSQHRPDKH
Summary
Uniprot
H9J766
A0A194QSH4
A0A0L7L3M8
A0A194PM76
A0A2H1VY68
A0A212EU10
+ More
A0A194QXY3 A0A2W1BE00 H9J767 A0A194PN60 A0A2H1V3K8 H9JJX6 A0A0L7LBN0 A0A2A4JSU1 A0A194RD92 B4MZT6 A0A1Q3FS04 B0X965 D6WK24 B4LUL9 A0A0L7L3I9 A0A182HAL3 A0A1S4FE70 B0X962 A0A182VSC8 A0A182LS85 A0A212EU23 A0A182YGJ3 D6WK25 E1ZZJ3 A0A1L8DZA3 A0A232ERT3 N6UGU5 Q175M8 K7J1Y2 D2A2D6 A0A1Q3FQ71 A0A182N5P4 K7J679 B4JEA0 A0A182VJF2 A0A182RYJ8 B4K144 Q175N0 A0A0J7KTS7 A0A2K6VB89 U4U5Y9 A0A194PQS6 A0A154PDS1 A0A1S4FE87 A0A182PF65 A0A182L7G6 A0A182XIZ5 B3MU60 K7J940 A0A0M9A434 D6WK26 A0A1S4FE25 Q175N1 A0A3B0KAR0 V5GP99 A0A2P8YLX0 Q7QKT9 A0A182TWI4 A0A088A736 Q7PQL5 K7IPV1 A0A2A3E710 V9IFX7 A0A1I8NRG7 A0A182I7W2 A0A232FKC4 A0A0P6J630 A0A1W4UV69 A0A182GQL3 B4MWY7 A0A182HAL4 A0A3B0JIA0 A0A3B0JIU6 E2BDR5 W8C7X0 A0A0Q9WRB1 A0A0K8UYI1 A0A1S4HCD0 A0A182WT65 A0A182K3L5 B4P705 F4WPC3
A0A194QXY3 A0A2W1BE00 H9J767 A0A194PN60 A0A2H1V3K8 H9JJX6 A0A0L7LBN0 A0A2A4JSU1 A0A194RD92 B4MZT6 A0A1Q3FS04 B0X965 D6WK24 B4LUL9 A0A0L7L3I9 A0A182HAL3 A0A1S4FE70 B0X962 A0A182VSC8 A0A182LS85 A0A212EU23 A0A182YGJ3 D6WK25 E1ZZJ3 A0A1L8DZA3 A0A232ERT3 N6UGU5 Q175M8 K7J1Y2 D2A2D6 A0A1Q3FQ71 A0A182N5P4 K7J679 B4JEA0 A0A182VJF2 A0A182RYJ8 B4K144 Q175N0 A0A0J7KTS7 A0A2K6VB89 U4U5Y9 A0A194PQS6 A0A154PDS1 A0A1S4FE87 A0A182PF65 A0A182L7G6 A0A182XIZ5 B3MU60 K7J940 A0A0M9A434 D6WK26 A0A1S4FE25 Q175N1 A0A3B0KAR0 V5GP99 A0A2P8YLX0 Q7QKT9 A0A182TWI4 A0A088A736 Q7PQL5 K7IPV1 A0A2A3E710 V9IFX7 A0A1I8NRG7 A0A182I7W2 A0A232FKC4 A0A0P6J630 A0A1W4UV69 A0A182GQL3 B4MWY7 A0A182HAL4 A0A3B0JIA0 A0A3B0JIU6 E2BDR5 W8C7X0 A0A0Q9WRB1 A0A0K8UYI1 A0A1S4HCD0 A0A182WT65 A0A182K3L5 B4P705 F4WPC3
Pubmed
EMBL
BABH01013774
KQ461154
KPJ08437.1
JTDY01003170
KOB70020.1
KQ459599
+ More
KPI94427.1 ODYU01005000 SOQ45442.1 AGBW02012491 OWR44992.1 KPJ08436.1 KZ150126 PZC73108.1 KPI94428.1 ODYU01000324 SOQ34864.1 BABH01026750 JTDY01001890 KOB72621.1 NWSH01000742 PCG74462.1 KQ460367 KPJ15439.1 CH963920 EDW77871.1 GFDL01004654 JAV30391.1 DS232520 EDS42971.1 KQ971342 EFA03937.1 CH940649 EDW64205.1 JTDY01003245 KOB69886.1 JXUM01122927 KQ566659 KXJ69933.1 EDS42968.1 AXCM01007881 OWR44993.1 EFA03627.2 GL435377 EFN73398.1 GFDF01002293 JAV11791.1 NNAY01002559 OXU21047.1 APGK01035612 KB740928 ENN77872.1 CH477398 EAT41774.1 KQ971338 EFA02038.1 GFDL01005328 JAV29717.1 CH916368 EDW03620.1 CH916740 EDW04553.1 EAT41772.1 LBMM01003342 KMQ93639.1 KB632064 ERL88482.1 KQ459595 KPI95801.1 KQ434869 KZC09408.1 CH902624 EDV33389.1 AAZX01018058 KQ435762 KOX75449.1 EFA03626.1 EAT41771.1 OUUW01000006 SPP82131.1 GALX01002542 JAB65924.1 PYGN01000502 PSN45249.1 AAAB01008441 EAA03387.4 AAAB01008880 EAA08644.6 KZ288353 PBC27284.1 JR045443 AEY59980.1 APCN01002561 NNAY01000087 OXU31125.1 GDUN01000271 JAN95648.1 JXUM01080854 KQ563212 KXJ74283.1 CH963857 EDW76626.1 KXJ69934.1 SPP82134.1 SPP82135.1 GL447689 EFN86121.1 GAMC01003359 JAC03197.1 KRF98755.1 GDHF01020924 JAI31390.1 AAAB01008964 CM000158 EDW89974.2 GL888243 EGI64018.1
KPI94427.1 ODYU01005000 SOQ45442.1 AGBW02012491 OWR44992.1 KPJ08436.1 KZ150126 PZC73108.1 KPI94428.1 ODYU01000324 SOQ34864.1 BABH01026750 JTDY01001890 KOB72621.1 NWSH01000742 PCG74462.1 KQ460367 KPJ15439.1 CH963920 EDW77871.1 GFDL01004654 JAV30391.1 DS232520 EDS42971.1 KQ971342 EFA03937.1 CH940649 EDW64205.1 JTDY01003245 KOB69886.1 JXUM01122927 KQ566659 KXJ69933.1 EDS42968.1 AXCM01007881 OWR44993.1 EFA03627.2 GL435377 EFN73398.1 GFDF01002293 JAV11791.1 NNAY01002559 OXU21047.1 APGK01035612 KB740928 ENN77872.1 CH477398 EAT41774.1 KQ971338 EFA02038.1 GFDL01005328 JAV29717.1 CH916368 EDW03620.1 CH916740 EDW04553.1 EAT41772.1 LBMM01003342 KMQ93639.1 KB632064 ERL88482.1 KQ459595 KPI95801.1 KQ434869 KZC09408.1 CH902624 EDV33389.1 AAZX01018058 KQ435762 KOX75449.1 EFA03626.1 EAT41771.1 OUUW01000006 SPP82131.1 GALX01002542 JAB65924.1 PYGN01000502 PSN45249.1 AAAB01008441 EAA03387.4 AAAB01008880 EAA08644.6 KZ288353 PBC27284.1 JR045443 AEY59980.1 APCN01002561 NNAY01000087 OXU31125.1 GDUN01000271 JAN95648.1 JXUM01080854 KQ563212 KXJ74283.1 CH963857 EDW76626.1 KXJ69934.1 SPP82134.1 SPP82135.1 GL447689 EFN86121.1 GAMC01003359 JAC03197.1 KRF98755.1 GDHF01020924 JAI31390.1 AAAB01008964 CM000158 EDW89974.2 GL888243 EGI64018.1
Proteomes
UP000005204
UP000053240
UP000037510
UP000053268
UP000007151
UP000218220
+ More
UP000007798 UP000002320 UP000007266 UP000008792 UP000069940 UP000249989 UP000075920 UP000075883 UP000076408 UP000000311 UP000215335 UP000019118 UP000008820 UP000002358 UP000075884 UP000001070 UP000075903 UP000075900 UP000036403 UP000030742 UP000076502 UP000075885 UP000075882 UP000076407 UP000007801 UP000053105 UP000268350 UP000245037 UP000007062 UP000075902 UP000005203 UP000242457 UP000095300 UP000075840 UP000192221 UP000008237 UP000075881 UP000002282 UP000007755
UP000007798 UP000002320 UP000007266 UP000008792 UP000069940 UP000249989 UP000075920 UP000075883 UP000076408 UP000000311 UP000215335 UP000019118 UP000008820 UP000002358 UP000075884 UP000001070 UP000075903 UP000075900 UP000036403 UP000030742 UP000076502 UP000075885 UP000075882 UP000076407 UP000007801 UP000053105 UP000268350 UP000245037 UP000007062 UP000075902 UP000005203 UP000242457 UP000095300 UP000075840 UP000192221 UP000008237 UP000075881 UP000002282 UP000007755
Interpro
Gene 3D
ProteinModelPortal
H9J766
A0A194QSH4
A0A0L7L3M8
A0A194PM76
A0A2H1VY68
A0A212EU10
+ More
A0A194QXY3 A0A2W1BE00 H9J767 A0A194PN60 A0A2H1V3K8 H9JJX6 A0A0L7LBN0 A0A2A4JSU1 A0A194RD92 B4MZT6 A0A1Q3FS04 B0X965 D6WK24 B4LUL9 A0A0L7L3I9 A0A182HAL3 A0A1S4FE70 B0X962 A0A182VSC8 A0A182LS85 A0A212EU23 A0A182YGJ3 D6WK25 E1ZZJ3 A0A1L8DZA3 A0A232ERT3 N6UGU5 Q175M8 K7J1Y2 D2A2D6 A0A1Q3FQ71 A0A182N5P4 K7J679 B4JEA0 A0A182VJF2 A0A182RYJ8 B4K144 Q175N0 A0A0J7KTS7 A0A2K6VB89 U4U5Y9 A0A194PQS6 A0A154PDS1 A0A1S4FE87 A0A182PF65 A0A182L7G6 A0A182XIZ5 B3MU60 K7J940 A0A0M9A434 D6WK26 A0A1S4FE25 Q175N1 A0A3B0KAR0 V5GP99 A0A2P8YLX0 Q7QKT9 A0A182TWI4 A0A088A736 Q7PQL5 K7IPV1 A0A2A3E710 V9IFX7 A0A1I8NRG7 A0A182I7W2 A0A232FKC4 A0A0P6J630 A0A1W4UV69 A0A182GQL3 B4MWY7 A0A182HAL4 A0A3B0JIA0 A0A3B0JIU6 E2BDR5 W8C7X0 A0A0Q9WRB1 A0A0K8UYI1 A0A1S4HCD0 A0A182WT65 A0A182K3L5 B4P705 F4WPC3
A0A194QXY3 A0A2W1BE00 H9J767 A0A194PN60 A0A2H1V3K8 H9JJX6 A0A0L7LBN0 A0A2A4JSU1 A0A194RD92 B4MZT6 A0A1Q3FS04 B0X965 D6WK24 B4LUL9 A0A0L7L3I9 A0A182HAL3 A0A1S4FE70 B0X962 A0A182VSC8 A0A182LS85 A0A212EU23 A0A182YGJ3 D6WK25 E1ZZJ3 A0A1L8DZA3 A0A232ERT3 N6UGU5 Q175M8 K7J1Y2 D2A2D6 A0A1Q3FQ71 A0A182N5P4 K7J679 B4JEA0 A0A182VJF2 A0A182RYJ8 B4K144 Q175N0 A0A0J7KTS7 A0A2K6VB89 U4U5Y9 A0A194PQS6 A0A154PDS1 A0A1S4FE87 A0A182PF65 A0A182L7G6 A0A182XIZ5 B3MU60 K7J940 A0A0M9A434 D6WK26 A0A1S4FE25 Q175N1 A0A3B0KAR0 V5GP99 A0A2P8YLX0 Q7QKT9 A0A182TWI4 A0A088A736 Q7PQL5 K7IPV1 A0A2A3E710 V9IFX7 A0A1I8NRG7 A0A182I7W2 A0A232FKC4 A0A0P6J630 A0A1W4UV69 A0A182GQL3 B4MWY7 A0A182HAL4 A0A3B0JIA0 A0A3B0JIU6 E2BDR5 W8C7X0 A0A0Q9WRB1 A0A0K8UYI1 A0A1S4HCD0 A0A182WT65 A0A182K3L5 B4P705 F4WPC3
Ontologies
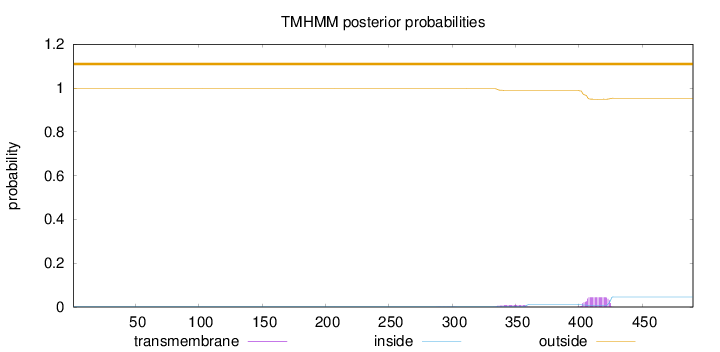
Topology
Length:
490
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.07993
Exp number, first 60 AAs:
0.00058
Total prob of N-in:
0.00302
outside
1 - 490
Population Genetic Test Statistics
Pi
162.21152
Theta
166.669349
Tajima's D
-0.506116
CLR
13.870833
CSRT
0.242637868106595
Interpretation
Uncertain
