Pre Gene Modal
BGIBMGA005451
Annotation
BET1-like_protein_[Bombyx_mori]
Full name
Adenylate kinase
Alternative Name
ATP-AMP transphosphorylase
ATP:AMP phosphotransferase
Adenylate kinase cytosolic and mitochondrial
Adenylate monophosphate kinase
ATP:AMP phosphotransferase
Adenylate kinase cytosolic and mitochondrial
Adenylate monophosphate kinase
Location in the cell
Cytoplasmic Reliability : 1.532
Sequence
CDS
ATGAGGCGAGCCCGCGAAGGCTACCACTATCAGCCAGTGCCCAGAGTAGCTACGGAAGATACTATCGAAAATGAAAATGAACGAATGGCTGAAGAACTCAGTGGCAAGATATCTTCACTTAAATACATTTCCATTGAGTTAGGCAATGAAGTAAGGGACCAAGAAAAACTTTTGCGTGGATTAGATGATGATGTTGACAGAAGCTCAGGTTTCCTTGGAAAAACCATGGGAAGAGTTTTAAGGCTTGGCAAAGGGAACCACAACTATTATATTTTCTATCTATTCCTCTTCTCCATTTTTGTGTTCTTTTTGTTGTATATTATTTTAAAATTTAGATGA
Protein
MRRAREGYHYQPVPRVATEDTIENENERMAEELSGKISSLKYISIELGNEVRDQEKLLRGLDDDVDRSSGFLGKTMGRVLRLGKGNHNYYIFYLFLFSIFVFFLLYIILKFR
Summary
Description
Catalyzes the reversible transfer of the terminal phosphate group between ATP and AMP. Plays an important role in cellular energy homeostasis and in adenine nucleotide metabolism. Adenylate kinase activity is critical for regulation of the phosphate utilization and the AMP de novo biosynthesis pathways.
Catalytic Activity
AMP + ATP = 2 ADP
Subunit
Monomer.
Similarity
Belongs to the adenylate kinase family. AK2 subfamily.
Uniprot
Q1HQ33
I4DJS2
A0A2H1VQR1
A0A0L7LUA2
A0A194RC87
A0A212EI83
+ More
D6X1L1 A0A1Y1N2J4 A0A2P8Z095 A0A0K8TNP3 E2BC20 A0A026VXK5 A0A151IQI7 A0A1Q3FDN7 A0A088ALI2 A0A2A3EA78 A0A154P2U4 A0A151ILZ6 A0A1B6KK99 A0A1B6GGZ1 A0A1B6HZV2 A0A1B6KR69 A0A1B6FH72 B0WTL9 E9JAW9 A0A0L7QMK1 A0A1I8NT41 A0A1B6JPK0 E2ALV2 U4U0L8 J3JWV7 A0A1Q3FD71 A0A2J7QFJ2 A0A0J7KR19 A0A0P4VUN7 R4G501 A0A151J7Q5 A0A158N934 A0A151X4Z9 F4WHG8 A0A2R7WYH8 W8CD23 A0A1B6CVQ8 A0A195FVN2 A0A1B6EFK4 T1GGW8 A0A067QNJ5 A0A195BTV2 K7J8L2 E0W482 A0A0C9QCI1 U5EW94 A0A182PBQ0 A0A182Y1U5 A0A182S8U1 A0A182VIJ9 A0A182UCI7 A0A182KSQ2 A0A182XD81 Q7Q060 A0A182HQ13 A0A1S3DT01 W5JNI7 A0A2L2Y9M6 A0A232ESH4 A0A084WHY3 A0A182JCK2 A0A1J1IT26 E2AEK4 A0A182M098 A0A182W977 A0A1L8DA37 A0A182QCC7 A0A182JNZ0 A0A182R5F7 A0A182MYI4 A0A1D2M4R4 A0A182FU43 A0A2M4C3T8 A0A336MV24 T1E7B0 A0A2M3ZAQ1 A0A1W4UQW1 A0A0M8ZYK6 A0A2L2Y960 A0A1L8E8Q1 T1IH28 A0A1L8ED46 A0A226DKQ0 A0A1A9Y0X2 B4ITM7 B4IG08 B4QQE0 B3NDY6 A0A087TV38 A0A0A9X851 A0A1I8ML06
D6X1L1 A0A1Y1N2J4 A0A2P8Z095 A0A0K8TNP3 E2BC20 A0A026VXK5 A0A151IQI7 A0A1Q3FDN7 A0A088ALI2 A0A2A3EA78 A0A154P2U4 A0A151ILZ6 A0A1B6KK99 A0A1B6GGZ1 A0A1B6HZV2 A0A1B6KR69 A0A1B6FH72 B0WTL9 E9JAW9 A0A0L7QMK1 A0A1I8NT41 A0A1B6JPK0 E2ALV2 U4U0L8 J3JWV7 A0A1Q3FD71 A0A2J7QFJ2 A0A0J7KR19 A0A0P4VUN7 R4G501 A0A151J7Q5 A0A158N934 A0A151X4Z9 F4WHG8 A0A2R7WYH8 W8CD23 A0A1B6CVQ8 A0A195FVN2 A0A1B6EFK4 T1GGW8 A0A067QNJ5 A0A195BTV2 K7J8L2 E0W482 A0A0C9QCI1 U5EW94 A0A182PBQ0 A0A182Y1U5 A0A182S8U1 A0A182VIJ9 A0A182UCI7 A0A182KSQ2 A0A182XD81 Q7Q060 A0A182HQ13 A0A1S3DT01 W5JNI7 A0A2L2Y9M6 A0A232ESH4 A0A084WHY3 A0A182JCK2 A0A1J1IT26 E2AEK4 A0A182M098 A0A182W977 A0A1L8DA37 A0A182QCC7 A0A182JNZ0 A0A182R5F7 A0A182MYI4 A0A1D2M4R4 A0A182FU43 A0A2M4C3T8 A0A336MV24 T1E7B0 A0A2M3ZAQ1 A0A1W4UQW1 A0A0M8ZYK6 A0A2L2Y960 A0A1L8E8Q1 T1IH28 A0A1L8ED46 A0A226DKQ0 A0A1A9Y0X2 B4ITM7 B4IG08 B4QQE0 B3NDY6 A0A087TV38 A0A0A9X851 A0A1I8ML06
EC Number
2.7.4.3
Pubmed
19121390
22651552
26354079
26227816
22118469
18362917
+ More
19820115 28004739 29403074 26369729 20798317 24508170 30249741 21282665 23537049 22516182 27129103 21347285 21719571 24495485 24845553 20075255 20566863 25244985 20966253 12364791 14747013 17210077 20920257 23761445 26561354 28648823 24438588 27289101 17994087 17550304 22936249 25401762 26823975 25315136
19820115 28004739 29403074 26369729 20798317 24508170 30249741 21282665 23537049 22516182 27129103 21347285 21719571 24495485 24845553 20075255 20566863 25244985 20966253 12364791 14747013 17210077 20920257 23761445 26561354 28648823 24438588 27289101 17994087 17550304 22936249 25401762 26823975 25315136
EMBL
BABH01013773
DQ443219
ABF51308.1
AK401540
KQ459599
BAM18162.1
+ More
KPI94474.1 ODYU01003876 SOQ43181.1 JTDY01000068 KOB79058.1 KQ460397 KPJ15217.1 AGBW02014681 OWR41196.1 KQ971370 EFA09340.1 GEZM01014307 JAV92143.1 PYGN01000257 PSN49925.1 GDAI01001845 JAI15758.1 GL447247 EFN86745.1 KK107638 QOIP01000004 EZA48405.1 RLU23692.1 KQ976785 KYN08303.1 GFDL01009368 JAV25677.1 KZ288324 PBC28086.1 KQ434807 KZC06152.1 KQ977085 KYN05915.1 GEBQ01028106 JAT11871.1 GECZ01008206 JAS61563.1 GECU01027514 GECU01006052 JAS80192.1 JAT01655.1 GEBQ01026025 JAT13952.1 GECZ01020207 JAS49562.1 DS232089 EDS34472.1 GL770679 EFZ10062.1 KQ414889 KOC59842.1 GECU01006520 JAT01187.1 GL440652 EFN65586.1 KB631843 ERL86632.1 APGK01044296 BT127725 KB741022 AEE62687.1 ENN74998.1 GFDL01009572 JAV25473.1 NEVH01015297 PNF27354.1 LBMM01004075 KMQ92827.1 GDKW01000177 JAI56418.1 ACPB03010353 GAHY01000925 JAA76585.1 KQ979640 KYN20048.1 ADTU01009059 KQ982543 KYQ55359.1 GL888161 EGI66322.1 KK856000 PTY24589.1 GAMC01002696 GAMC01002694 JAC03860.1 GEDC01019669 JAS17629.1 KQ981268 KYN43919.1 GEDC01000628 JAS36670.1 CAQQ02178081 KK853197 KDR09963.1 KQ976405 KYM91864.1 AAZX01007490 DS235886 EEB20438.1 GBYB01001009 JAG70776.1 GANO01000666 JAB59205.1 AAAB01008986 EAA00304.2 APCN01003288 ADMH02001044 ETN64339.1 IAAA01012538 IAAA01012539 LAA04697.1 NNAY01002421 OXU21310.1 ATLV01023896 KE525347 KFB49827.1 AXCP01006759 CVRI01000056 CRL01657.1 GL438870 EFN68127.1 AXCM01002401 GFDF01010900 JAV03184.1 AXCN02001041 LJIJ01004309 ODM87968.1 GGFJ01010794 MBW59935.1 UFQT01001890 SSX32097.1 GAMD01003197 JAA98393.1 GGFM01004820 MBW25571.1 KQ435794 KOX73740.1 IAAA01012537 LAA04688.1 GFDG01003755 JAV15044.1 JH429682 GFDG01002148 JAV16651.1 LNIX01000017 OXA45700.1 CM000159 CH891721 EDW95351.1 EDW99740.1 CH480834 EDW46595.1 CM000363 CM002912 EDX11046.1 KMZ00509.1 CH954178 EDV52340.1 KK116875 KFM68977.1 GBHO01027783 GBRD01000468 GDHC01008544 JAG15821.1 JAG65353.1 JAQ10085.1
KPI94474.1 ODYU01003876 SOQ43181.1 JTDY01000068 KOB79058.1 KQ460397 KPJ15217.1 AGBW02014681 OWR41196.1 KQ971370 EFA09340.1 GEZM01014307 JAV92143.1 PYGN01000257 PSN49925.1 GDAI01001845 JAI15758.1 GL447247 EFN86745.1 KK107638 QOIP01000004 EZA48405.1 RLU23692.1 KQ976785 KYN08303.1 GFDL01009368 JAV25677.1 KZ288324 PBC28086.1 KQ434807 KZC06152.1 KQ977085 KYN05915.1 GEBQ01028106 JAT11871.1 GECZ01008206 JAS61563.1 GECU01027514 GECU01006052 JAS80192.1 JAT01655.1 GEBQ01026025 JAT13952.1 GECZ01020207 JAS49562.1 DS232089 EDS34472.1 GL770679 EFZ10062.1 KQ414889 KOC59842.1 GECU01006520 JAT01187.1 GL440652 EFN65586.1 KB631843 ERL86632.1 APGK01044296 BT127725 KB741022 AEE62687.1 ENN74998.1 GFDL01009572 JAV25473.1 NEVH01015297 PNF27354.1 LBMM01004075 KMQ92827.1 GDKW01000177 JAI56418.1 ACPB03010353 GAHY01000925 JAA76585.1 KQ979640 KYN20048.1 ADTU01009059 KQ982543 KYQ55359.1 GL888161 EGI66322.1 KK856000 PTY24589.1 GAMC01002696 GAMC01002694 JAC03860.1 GEDC01019669 JAS17629.1 KQ981268 KYN43919.1 GEDC01000628 JAS36670.1 CAQQ02178081 KK853197 KDR09963.1 KQ976405 KYM91864.1 AAZX01007490 DS235886 EEB20438.1 GBYB01001009 JAG70776.1 GANO01000666 JAB59205.1 AAAB01008986 EAA00304.2 APCN01003288 ADMH02001044 ETN64339.1 IAAA01012538 IAAA01012539 LAA04697.1 NNAY01002421 OXU21310.1 ATLV01023896 KE525347 KFB49827.1 AXCP01006759 CVRI01000056 CRL01657.1 GL438870 EFN68127.1 AXCM01002401 GFDF01010900 JAV03184.1 AXCN02001041 LJIJ01004309 ODM87968.1 GGFJ01010794 MBW59935.1 UFQT01001890 SSX32097.1 GAMD01003197 JAA98393.1 GGFM01004820 MBW25571.1 KQ435794 KOX73740.1 IAAA01012537 LAA04688.1 GFDG01003755 JAV15044.1 JH429682 GFDG01002148 JAV16651.1 LNIX01000017 OXA45700.1 CM000159 CH891721 EDW95351.1 EDW99740.1 CH480834 EDW46595.1 CM000363 CM002912 EDX11046.1 KMZ00509.1 CH954178 EDV52340.1 KK116875 KFM68977.1 GBHO01027783 GBRD01000468 GDHC01008544 JAG15821.1 JAG65353.1 JAQ10085.1
Proteomes
UP000005204
UP000053268
UP000037510
UP000053240
UP000007151
UP000007266
+ More
UP000245037 UP000008237 UP000053097 UP000279307 UP000078542 UP000005203 UP000242457 UP000076502 UP000002320 UP000053825 UP000095300 UP000000311 UP000030742 UP000019118 UP000235965 UP000036403 UP000015103 UP000078492 UP000005205 UP000075809 UP000007755 UP000078541 UP000015102 UP000027135 UP000078540 UP000002358 UP000009046 UP000075885 UP000076408 UP000075901 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000079169 UP000000673 UP000215335 UP000030765 UP000075880 UP000183832 UP000075883 UP000075920 UP000075886 UP000075881 UP000075900 UP000075884 UP000094527 UP000069272 UP000192221 UP000053105 UP000198287 UP000092443 UP000002282 UP000001292 UP000000304 UP000008711 UP000054359 UP000095301
UP000245037 UP000008237 UP000053097 UP000279307 UP000078542 UP000005203 UP000242457 UP000076502 UP000002320 UP000053825 UP000095300 UP000000311 UP000030742 UP000019118 UP000235965 UP000036403 UP000015103 UP000078492 UP000005205 UP000075809 UP000007755 UP000078541 UP000015102 UP000027135 UP000078540 UP000002358 UP000009046 UP000075885 UP000076408 UP000075901 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000079169 UP000000673 UP000215335 UP000030765 UP000075880 UP000183832 UP000075883 UP000075920 UP000075886 UP000075881 UP000075900 UP000075884 UP000094527 UP000069272 UP000192221 UP000053105 UP000198287 UP000092443 UP000002282 UP000001292 UP000000304 UP000008711 UP000054359 UP000095301
PRIDE
Interpro
IPR039897
BET1
+ More
IPR039899 BET1_SNARE
IPR000727 T_SNARE_dom
IPR005606 Sec20
IPR027417 P-loop_NTPase
IPR033690 Adenylat_kinase_CS
IPR007862 Adenylate_kinase_lid-dom
IPR000850 Adenylat/UMP-CMP_kin
IPR006259 Adenyl_kin_sub
IPR028587 AK2
IPR020683 Ankyrin_rpt-contain_dom
IPR021939 KN_motif
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
IPR039899 BET1_SNARE
IPR000727 T_SNARE_dom
IPR005606 Sec20
IPR027417 P-loop_NTPase
IPR033690 Adenylat_kinase_CS
IPR007862 Adenylate_kinase_lid-dom
IPR000850 Adenylat/UMP-CMP_kin
IPR006259 Adenyl_kin_sub
IPR028587 AK2
IPR020683 Ankyrin_rpt-contain_dom
IPR021939 KN_motif
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
Gene 3D
ProteinModelPortal
Q1HQ33
I4DJS2
A0A2H1VQR1
A0A0L7LUA2
A0A194RC87
A0A212EI83
+ More
D6X1L1 A0A1Y1N2J4 A0A2P8Z095 A0A0K8TNP3 E2BC20 A0A026VXK5 A0A151IQI7 A0A1Q3FDN7 A0A088ALI2 A0A2A3EA78 A0A154P2U4 A0A151ILZ6 A0A1B6KK99 A0A1B6GGZ1 A0A1B6HZV2 A0A1B6KR69 A0A1B6FH72 B0WTL9 E9JAW9 A0A0L7QMK1 A0A1I8NT41 A0A1B6JPK0 E2ALV2 U4U0L8 J3JWV7 A0A1Q3FD71 A0A2J7QFJ2 A0A0J7KR19 A0A0P4VUN7 R4G501 A0A151J7Q5 A0A158N934 A0A151X4Z9 F4WHG8 A0A2R7WYH8 W8CD23 A0A1B6CVQ8 A0A195FVN2 A0A1B6EFK4 T1GGW8 A0A067QNJ5 A0A195BTV2 K7J8L2 E0W482 A0A0C9QCI1 U5EW94 A0A182PBQ0 A0A182Y1U5 A0A182S8U1 A0A182VIJ9 A0A182UCI7 A0A182KSQ2 A0A182XD81 Q7Q060 A0A182HQ13 A0A1S3DT01 W5JNI7 A0A2L2Y9M6 A0A232ESH4 A0A084WHY3 A0A182JCK2 A0A1J1IT26 E2AEK4 A0A182M098 A0A182W977 A0A1L8DA37 A0A182QCC7 A0A182JNZ0 A0A182R5F7 A0A182MYI4 A0A1D2M4R4 A0A182FU43 A0A2M4C3T8 A0A336MV24 T1E7B0 A0A2M3ZAQ1 A0A1W4UQW1 A0A0M8ZYK6 A0A2L2Y960 A0A1L8E8Q1 T1IH28 A0A1L8ED46 A0A226DKQ0 A0A1A9Y0X2 B4ITM7 B4IG08 B4QQE0 B3NDY6 A0A087TV38 A0A0A9X851 A0A1I8ML06
D6X1L1 A0A1Y1N2J4 A0A2P8Z095 A0A0K8TNP3 E2BC20 A0A026VXK5 A0A151IQI7 A0A1Q3FDN7 A0A088ALI2 A0A2A3EA78 A0A154P2U4 A0A151ILZ6 A0A1B6KK99 A0A1B6GGZ1 A0A1B6HZV2 A0A1B6KR69 A0A1B6FH72 B0WTL9 E9JAW9 A0A0L7QMK1 A0A1I8NT41 A0A1B6JPK0 E2ALV2 U4U0L8 J3JWV7 A0A1Q3FD71 A0A2J7QFJ2 A0A0J7KR19 A0A0P4VUN7 R4G501 A0A151J7Q5 A0A158N934 A0A151X4Z9 F4WHG8 A0A2R7WYH8 W8CD23 A0A1B6CVQ8 A0A195FVN2 A0A1B6EFK4 T1GGW8 A0A067QNJ5 A0A195BTV2 K7J8L2 E0W482 A0A0C9QCI1 U5EW94 A0A182PBQ0 A0A182Y1U5 A0A182S8U1 A0A182VIJ9 A0A182UCI7 A0A182KSQ2 A0A182XD81 Q7Q060 A0A182HQ13 A0A1S3DT01 W5JNI7 A0A2L2Y9M6 A0A232ESH4 A0A084WHY3 A0A182JCK2 A0A1J1IT26 E2AEK4 A0A182M098 A0A182W977 A0A1L8DA37 A0A182QCC7 A0A182JNZ0 A0A182R5F7 A0A182MYI4 A0A1D2M4R4 A0A182FU43 A0A2M4C3T8 A0A336MV24 T1E7B0 A0A2M3ZAQ1 A0A1W4UQW1 A0A0M8ZYK6 A0A2L2Y960 A0A1L8E8Q1 T1IH28 A0A1L8ED46 A0A226DKQ0 A0A1A9Y0X2 B4ITM7 B4IG08 B4QQE0 B3NDY6 A0A087TV38 A0A0A9X851 A0A1I8ML06
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Predominantly mitochondrial. With evidence from 1 publications.
Cytosol Predominantly mitochondrial. With evidence from 1 publications.
Mitochondrion intermembrane space Predominantly mitochondrial. With evidence from 1 publications.
Cytosol Predominantly mitochondrial. With evidence from 1 publications.
Mitochondrion intermembrane space Predominantly mitochondrial. With evidence from 1 publications.
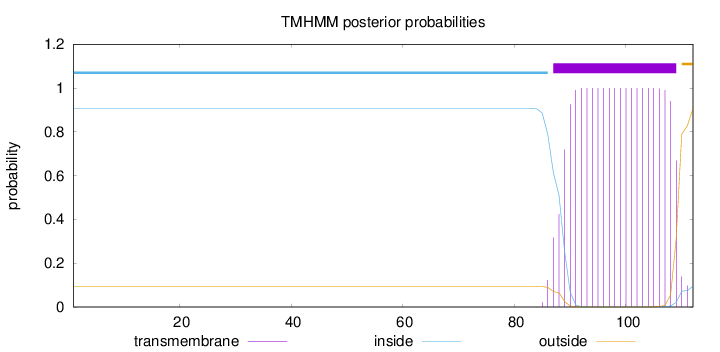
Length:
112
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.35259
Exp number, first 60 AAs:
0
Total prob of N-in:
0.90587
inside
1 - 86
TMhelix
87 - 109
outside
110 - 112
Population Genetic Test Statistics
Pi
78.060326
Theta
136.118406
Tajima's D
-1.122508
CLR
314.01962
CSRT
0.115144242787861
Interpretation
Uncertain
