Gene
KWMTBOMO04506
Pre Gene Modal
BGIBMGA005360
Annotation
PREDICTED:_conserved_oligomeric_Golgi_complex_subunit_8_isoform_X2_[Bombyx_mori]
Full name
Conserved oligomeric Golgi complex subunit 8
Alternative Name
Component of oligomeric Golgi complex 8
Location in the cell
Nuclear Reliability : 3.746
Sequence
CDS
ATGTCTCAGGAACTAAAGGAACTCTGTCAATTAATTTTCCCAAGCTCACCTACAGAAAATGTCATATTTTTTAGCGAAGTCAGTGATTACATCAAAAAATTGGGTTCTCAGAATTGGGACTATATTCGGAAAGAACCAGAAAGGCTTACTGAGGAGATGAAGCACTTAACAGAGCAGACCCAGGAATTAGCATTCACAAATTACAAGACTTTTGTTGAAACTGCAGAAATATCCCGTACTATAATTAAGGATCTTAATAGATCAAAGCAATCCTTACAGAATTTTCTTGATGCTACTCCTGGTTTTATTGAAGAGAGCGAAAGGTTTTCGCAGATGGCTGGTACCATTGTGAAAGAGAAAAGTCGATATAGCACTATCAGAAATCAAAGTGATAAGTTGTTAGAACTTCTTGAACTGCCGTCGTTGATGCGAGAGGCTTTAAATGCTGAAGATTATGAAAGTGCATTAGATATATTCACTTTTGTTCGAAATTTAAGTAAAAGGTACTCTGTGATACCAATTGTTCAGAATACAACAAATGAGATAATGACATTATGGTTTGAAACACTGTATCATCTCTTCAATCAGCTTCGTTATGATTTACCGCTTCCTCAGTGCCTACAAATATTAGGTTACCTGCGCAGAGCAAACACAGTTTACAAATCAACAGATGAAGAAGAAAACAGTATGCAGTCTATAATAGGGAACAAAAATACAAGTTCTGATGGATTACACTTACATTTCTTAAAAGCGAGAAATGCTTGGTTTGAAAAAGCTTTAGTTGATGCTAGAAATTCTGAATCATCTGATAAAGTTCTAAGAAGAATTGTGGAATTGCATAGAATACATCTCTTCAATGTATTAACCCAGCATAAATCAATTTTCTTATCTGATGCTCAAGAATCCAAAGCAAGAGATGATGAATTGAGTGGAAACAGTGCTCTATCAAACTGGTTAAAATTAAAGGTGGAGATTTTCGCGCATATTTTAAACCAAAATTTGGGTAAAGAAGATGAAGCTAATTTCGAATCACTTTTGAACCAATGTATGTACCTTTGTCTTTCGTTCGGAAGAGTCGGCGCTGACATGCGGTGTGTCTTAACGCCGCTGTTTCGCGACTGCATACTAAAGCAATTCCATTCAAGTCTTGATAAAGTCAGTGATCACTTTGAGCAACAAATGCGTGCATACAAAGTTCCTAGTATCAAAAACTATCCAAGACCAATTACGGAAACGGCAAAGGGTCCTCCCGAAAACCTCCTTGATTACTATCCACTAGCAGAGTACTGCAATGGAATGCTTACAGTCTTGAATTCTTTAAGAGTTACAGCTCCATTAAACATAGCTAAAGAAGTGTATAACTCATATAAAGAATCATTAAACAGTACAGTCCAAGTATTGTTAAATTTCTATTACAGAGAACAACAAGGATTTACTGATGTGGAGAGACAAAATTACGTATCATTGTGTCTTTCCTTTAGAGATGATTTAATACCTTACATCACTAAATGTTTATCTCAATCGTTCCCTTCAACACAAATAGCAGAGTTGCTAGGTGTTACTTTGACTGTTCTTCAAGACAGTAAAATTTTGTACATAGACCTCCATGCAATGTGTGAACCTTTAAATGTCATTACTGGTTCAAATTAA
Protein
MSQELKELCQLIFPSSPTENVIFFSEVSDYIKKLGSQNWDYIRKEPERLTEEMKHLTEQTQELAFTNYKTFVETAEISRTIIKDLNRSKQSLQNFLDATPGFIEESERFSQMAGTIVKEKSRYSTIRNQSDKLLELLELPSLMREALNAEDYESALDIFTFVRNLSKRYSVIPIVQNTTNEIMTLWFETLYHLFNQLRYDLPLPQCLQILGYLRRANTVYKSTDEEENSMQSIIGNKNTSSDGLHLHFLKARNAWFEKALVDARNSESSDKVLRRIVELHRIHLFNVLTQHKSIFLSDAQESKARDDELSGNSALSNWLKLKVEIFAHILNQNLGKEDEANFESLLNQCMYLCLSFGRVGADMRCVLTPLFRDCILKQFHSSLDKVSDHFEQQMRAYKVPSIKNYPRPITETAKGPPENLLDYYPLAEYCNGMLTVLNSLRVTAPLNIAKEVYNSYKESLNSTVQVLLNFYYREQQGFTDVERQNYVSLCLSFRDDLIPYITKCLSQSFPSTQIAELLGVTLTVLQDSKILYIDLHAMCEPLNVITGSN
Summary
Description
Required for normal Golgi function.
Subunit
Component of the conserved oligomeric Golgi complex which is composed of eight different subunits and is required for normal Golgi morphology and localization.
Similarity
Belongs to the COG8 family.
Keywords
Complete proteome
Golgi apparatus
Membrane
Protein transport
Reference proteome
Transport
Congenital disorder of glycosylation
Polymorphism
Feature
chain Conserved oligomeric Golgi complex subunit 8
sequence variant In dbSNP:rs3027.
sequence variant In dbSNP:rs3027.
Uniprot
H9J768
A0A2A4J7K6
A0A2H1X0R0
A0A1E1WUV8
A0A3S2NML7
S4PWY0
+ More
A0A212EI89 A0A194PMC5 A0A194RD62 A0A2J7R8N2 A0A067RM63 A0A2P8ZLY9 A0A1B6M794 A0A034V2J5 A0A0A1WG88 A0A1I8P4J1 A0A0K8U1H9 A0A0L0CGJ9 A0A1L8DVG2 B4LQF0 A0A1A9VXL8 A0A1I8ME71 A0A1B6DKQ7 A0A1A9XDS3 A0A1B0B0E4 A0A1B0G4H8 B3N4I1 A0A1A9ZIU5 A0A023EX81 B4G6P5 Q29M23 B4NMV6 A0A182GTS9 A0A1S4FK99 Q16XW7 B4Q370 B4HX53 B4P1G9 Q9VKH0 A0A1A9W1R1 K7J0M6 A0A1W4W055 A0A232FK12 E2A4G1 W8BKN4 B4JPH8 B3MMU4 A0A0M4EFP9 A0A3B0JAZ3 B0WGX3 B4KH87 N6UFE8 A0A1Q3F7X4 A0A084WBA5 A0A195D9R9 F4WXK7 E2BLS9 A0A026W2Z3 D2A1E2 A0A0J9R0C9 A0A154P8Y3 A0A195BXF7 U4UM63 A0A182R8M4 A0A182QH74 A0A1Q3F766 A0A2M4ACF7 Q7Q351 A0A182UE16 A0A2M4AC06 A0A182LPE9 A0A182IWC6 A0A182XC74 A0A182IIF8 A0A182USF3 A0A182K7Q1 A0A195CQJ0 A0A2M4CQL7 W5JT40 A0A182MMK2 A0A182SKE4 A0A182MY14 A0A2M3Z5K9 A0A2M3Z6C7 A0A0T6B415 A0A1Y1KQR1 A0A3F2YWK0 A0A310SS82 A0A182FKQ1 A0A1U7QZJ1 A0A2M4BHX4 A0A2M4BHE3 A0A024R6Z6 Q96MW5 A0A0N0BI72 U4UBE7 K7C7D5 A0A2R8Z9X5 K7CR24
A0A212EI89 A0A194PMC5 A0A194RD62 A0A2J7R8N2 A0A067RM63 A0A2P8ZLY9 A0A1B6M794 A0A034V2J5 A0A0A1WG88 A0A1I8P4J1 A0A0K8U1H9 A0A0L0CGJ9 A0A1L8DVG2 B4LQF0 A0A1A9VXL8 A0A1I8ME71 A0A1B6DKQ7 A0A1A9XDS3 A0A1B0B0E4 A0A1B0G4H8 B3N4I1 A0A1A9ZIU5 A0A023EX81 B4G6P5 Q29M23 B4NMV6 A0A182GTS9 A0A1S4FK99 Q16XW7 B4Q370 B4HX53 B4P1G9 Q9VKH0 A0A1A9W1R1 K7J0M6 A0A1W4W055 A0A232FK12 E2A4G1 W8BKN4 B4JPH8 B3MMU4 A0A0M4EFP9 A0A3B0JAZ3 B0WGX3 B4KH87 N6UFE8 A0A1Q3F7X4 A0A084WBA5 A0A195D9R9 F4WXK7 E2BLS9 A0A026W2Z3 D2A1E2 A0A0J9R0C9 A0A154P8Y3 A0A195BXF7 U4UM63 A0A182R8M4 A0A182QH74 A0A1Q3F766 A0A2M4ACF7 Q7Q351 A0A182UE16 A0A2M4AC06 A0A182LPE9 A0A182IWC6 A0A182XC74 A0A182IIF8 A0A182USF3 A0A182K7Q1 A0A195CQJ0 A0A2M4CQL7 W5JT40 A0A182MMK2 A0A182SKE4 A0A182MY14 A0A2M3Z5K9 A0A2M3Z6C7 A0A0T6B415 A0A1Y1KQR1 A0A3F2YWK0 A0A310SS82 A0A182FKQ1 A0A1U7QZJ1 A0A2M4BHX4 A0A2M4BHE3 A0A024R6Z6 Q96MW5 A0A0N0BI72 U4UBE7 K7C7D5 A0A2R8Z9X5 K7CR24
Pubmed
19121390
23622113
22118469
26354079
24845553
29403074
+ More
25348373 25830018 26108605 17994087 25315136 24945155 15632085 26483478 17510324 17550304 10731132 12537572 12537569 20075255 28648823 20798317 24495485 23537049 24438588 21719571 24508170 30249741 18362917 19820115 22936249 12364791 14747013 17210077 20966253 20920257 23761445 28004739 28071753 11181995 14702039 15489334 11703943 17331980 21269460 22722832 16136131
25348373 25830018 26108605 17994087 25315136 24945155 15632085 26483478 17510324 17550304 10731132 12537572 12537569 20075255 28648823 20798317 24495485 23537049 24438588 21719571 24508170 30249741 18362917 19820115 22936249 12364791 14747013 17210077 20966253 20920257 23761445 28004739 28071753 11181995 14702039 15489334 11703943 17331980 21269460 22722832 16136131
EMBL
BABH01013772
NWSH01002530
PCG68065.1
ODYU01012497
SOQ58838.1
GDQN01000286
+ More
JAT90768.1 RSAL01000032 RVE51520.1 GAIX01006003 JAA86557.1 AGBW02014681 OWR41198.1 KQ459599 KPI94472.1 KQ460397 KPJ15215.1 NEVH01006721 PNF37192.1 KK852543 KDR21660.1 PYGN01000019 PSN57517.1 GEBQ01008206 JAT31771.1 GAKP01022256 GAKP01022253 JAC36699.1 GBXI01016248 JAC98043.1 GDHF01031941 JAI20373.1 JRES01000433 KNC31337.1 GFDF01003668 JAV10416.1 CH940649 EDW63400.1 GEDC01011041 JAS26257.1 JXJN01006750 CCAG010000911 CH954177 EDV58893.1 GAPW01000604 JAC12994.1 CH479180 EDW29159.1 CH379060 EAL33872.3 CH964282 EDW85695.2 JXUM01087786 KQ563658 KXJ73537.1 CH477530 EAT39477.1 CM000361 EDX04701.1 CH480818 EDW52598.1 CM000157 EDW88076.1 AE014134 BT031276 AY069825 NNAY01000105 OXU30923.1 GL436635 EFN71708.1 GAMC01009022 JAB97533.1 CH916372 EDV98808.1 CH902620 EDV30969.1 CP012523 ALC39317.1 OUUW01000004 SPP79517.1 DS231930 EDS27267.1 CH933807 EDW13304.1 APGK01029087 KB740724 ENN79356.1 GFDL01011457 JAV23588.1 ATLV01022328 KE525331 KFB47499.1 KQ981082 KYN09612.1 GL888425 EGI61077.1 GL449036 EFN83413.1 KK107519 QOIP01000009 EZA49404.1 RLU18524.1 KQ971338 EFA02659.1 CM002910 KMY89757.1 KQ434829 KZC07678.1 KQ976401 KYM92633.1 KB632278 ERL91256.1 AXCN02001643 GFDL01011634 JAV23411.1 GGFK01005153 MBW38474.1 AAAB01008966 EAA13024.4 GGFK01004969 MBW38290.1 APCN01001931 KQ977444 KYN02742.1 GGFL01003448 MBW67626.1 ADMH02000526 ETN66110.1 AXCM01007124 GGFM01003051 MBW23802.1 GGFM01003321 MBW24072.1 LJIG01016010 KRT81886.1 GEZM01078375 JAV62751.1 KQ760645 OAD59682.1 GGFJ01003267 MBW52408.1 GGFJ01003272 MBW52413.1 CH471092 EAW83267.1 AK056344 AK025968 BC017492 BC121022 BC121023 KQ435733 KOX77238.1 ERL91254.1 GABE01006263 JAA38476.1 AJFE02047230 AJFE02047231 AJFE02047232 AACZ04051800 GABC01006188 GABD01006415 JAA05150.1 JAA26685.1
JAT90768.1 RSAL01000032 RVE51520.1 GAIX01006003 JAA86557.1 AGBW02014681 OWR41198.1 KQ459599 KPI94472.1 KQ460397 KPJ15215.1 NEVH01006721 PNF37192.1 KK852543 KDR21660.1 PYGN01000019 PSN57517.1 GEBQ01008206 JAT31771.1 GAKP01022256 GAKP01022253 JAC36699.1 GBXI01016248 JAC98043.1 GDHF01031941 JAI20373.1 JRES01000433 KNC31337.1 GFDF01003668 JAV10416.1 CH940649 EDW63400.1 GEDC01011041 JAS26257.1 JXJN01006750 CCAG010000911 CH954177 EDV58893.1 GAPW01000604 JAC12994.1 CH479180 EDW29159.1 CH379060 EAL33872.3 CH964282 EDW85695.2 JXUM01087786 KQ563658 KXJ73537.1 CH477530 EAT39477.1 CM000361 EDX04701.1 CH480818 EDW52598.1 CM000157 EDW88076.1 AE014134 BT031276 AY069825 NNAY01000105 OXU30923.1 GL436635 EFN71708.1 GAMC01009022 JAB97533.1 CH916372 EDV98808.1 CH902620 EDV30969.1 CP012523 ALC39317.1 OUUW01000004 SPP79517.1 DS231930 EDS27267.1 CH933807 EDW13304.1 APGK01029087 KB740724 ENN79356.1 GFDL01011457 JAV23588.1 ATLV01022328 KE525331 KFB47499.1 KQ981082 KYN09612.1 GL888425 EGI61077.1 GL449036 EFN83413.1 KK107519 QOIP01000009 EZA49404.1 RLU18524.1 KQ971338 EFA02659.1 CM002910 KMY89757.1 KQ434829 KZC07678.1 KQ976401 KYM92633.1 KB632278 ERL91256.1 AXCN02001643 GFDL01011634 JAV23411.1 GGFK01005153 MBW38474.1 AAAB01008966 EAA13024.4 GGFK01004969 MBW38290.1 APCN01001931 KQ977444 KYN02742.1 GGFL01003448 MBW67626.1 ADMH02000526 ETN66110.1 AXCM01007124 GGFM01003051 MBW23802.1 GGFM01003321 MBW24072.1 LJIG01016010 KRT81886.1 GEZM01078375 JAV62751.1 KQ760645 OAD59682.1 GGFJ01003267 MBW52408.1 GGFJ01003272 MBW52413.1 CH471092 EAW83267.1 AK056344 AK025968 BC017492 BC121022 BC121023 KQ435733 KOX77238.1 ERL91254.1 GABE01006263 JAA38476.1 AJFE02047230 AJFE02047231 AJFE02047232 AACZ04051800 GABC01006188 GABD01006415 JAA05150.1 JAA26685.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053268
UP000053240
+ More
UP000235965 UP000027135 UP000245037 UP000095300 UP000037069 UP000008792 UP000078200 UP000095301 UP000092443 UP000092460 UP000092444 UP000008711 UP000092445 UP000008744 UP000001819 UP000007798 UP000069940 UP000249989 UP000008820 UP000000304 UP000001292 UP000002282 UP000000803 UP000091820 UP000002358 UP000192221 UP000215335 UP000000311 UP000001070 UP000007801 UP000092553 UP000268350 UP000002320 UP000009192 UP000019118 UP000030765 UP000078492 UP000007755 UP000008237 UP000053097 UP000279307 UP000007266 UP000076502 UP000078540 UP000030742 UP000075900 UP000075886 UP000007062 UP000075902 UP000075882 UP000075880 UP000076407 UP000075840 UP000075903 UP000075881 UP000078542 UP000000673 UP000075883 UP000075901 UP000075884 UP000075885 UP000069272 UP000189706 UP000005640 UP000053105 UP000240080 UP000002277
UP000235965 UP000027135 UP000245037 UP000095300 UP000037069 UP000008792 UP000078200 UP000095301 UP000092443 UP000092460 UP000092444 UP000008711 UP000092445 UP000008744 UP000001819 UP000007798 UP000069940 UP000249989 UP000008820 UP000000304 UP000001292 UP000002282 UP000000803 UP000091820 UP000002358 UP000192221 UP000215335 UP000000311 UP000001070 UP000007801 UP000092553 UP000268350 UP000002320 UP000009192 UP000019118 UP000030765 UP000078492 UP000007755 UP000008237 UP000053097 UP000279307 UP000007266 UP000076502 UP000078540 UP000030742 UP000075900 UP000075886 UP000007062 UP000075902 UP000075882 UP000075880 UP000076407 UP000075840 UP000075903 UP000075881 UP000078542 UP000000673 UP000075883 UP000075901 UP000075884 UP000075885 UP000069272 UP000189706 UP000005640 UP000053105 UP000240080 UP000002277
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J768
A0A2A4J7K6
A0A2H1X0R0
A0A1E1WUV8
A0A3S2NML7
S4PWY0
+ More
A0A212EI89 A0A194PMC5 A0A194RD62 A0A2J7R8N2 A0A067RM63 A0A2P8ZLY9 A0A1B6M794 A0A034V2J5 A0A0A1WG88 A0A1I8P4J1 A0A0K8U1H9 A0A0L0CGJ9 A0A1L8DVG2 B4LQF0 A0A1A9VXL8 A0A1I8ME71 A0A1B6DKQ7 A0A1A9XDS3 A0A1B0B0E4 A0A1B0G4H8 B3N4I1 A0A1A9ZIU5 A0A023EX81 B4G6P5 Q29M23 B4NMV6 A0A182GTS9 A0A1S4FK99 Q16XW7 B4Q370 B4HX53 B4P1G9 Q9VKH0 A0A1A9W1R1 K7J0M6 A0A1W4W055 A0A232FK12 E2A4G1 W8BKN4 B4JPH8 B3MMU4 A0A0M4EFP9 A0A3B0JAZ3 B0WGX3 B4KH87 N6UFE8 A0A1Q3F7X4 A0A084WBA5 A0A195D9R9 F4WXK7 E2BLS9 A0A026W2Z3 D2A1E2 A0A0J9R0C9 A0A154P8Y3 A0A195BXF7 U4UM63 A0A182R8M4 A0A182QH74 A0A1Q3F766 A0A2M4ACF7 Q7Q351 A0A182UE16 A0A2M4AC06 A0A182LPE9 A0A182IWC6 A0A182XC74 A0A182IIF8 A0A182USF3 A0A182K7Q1 A0A195CQJ0 A0A2M4CQL7 W5JT40 A0A182MMK2 A0A182SKE4 A0A182MY14 A0A2M3Z5K9 A0A2M3Z6C7 A0A0T6B415 A0A1Y1KQR1 A0A3F2YWK0 A0A310SS82 A0A182FKQ1 A0A1U7QZJ1 A0A2M4BHX4 A0A2M4BHE3 A0A024R6Z6 Q96MW5 A0A0N0BI72 U4UBE7 K7C7D5 A0A2R8Z9X5 K7CR24
A0A212EI89 A0A194PMC5 A0A194RD62 A0A2J7R8N2 A0A067RM63 A0A2P8ZLY9 A0A1B6M794 A0A034V2J5 A0A0A1WG88 A0A1I8P4J1 A0A0K8U1H9 A0A0L0CGJ9 A0A1L8DVG2 B4LQF0 A0A1A9VXL8 A0A1I8ME71 A0A1B6DKQ7 A0A1A9XDS3 A0A1B0B0E4 A0A1B0G4H8 B3N4I1 A0A1A9ZIU5 A0A023EX81 B4G6P5 Q29M23 B4NMV6 A0A182GTS9 A0A1S4FK99 Q16XW7 B4Q370 B4HX53 B4P1G9 Q9VKH0 A0A1A9W1R1 K7J0M6 A0A1W4W055 A0A232FK12 E2A4G1 W8BKN4 B4JPH8 B3MMU4 A0A0M4EFP9 A0A3B0JAZ3 B0WGX3 B4KH87 N6UFE8 A0A1Q3F7X4 A0A084WBA5 A0A195D9R9 F4WXK7 E2BLS9 A0A026W2Z3 D2A1E2 A0A0J9R0C9 A0A154P8Y3 A0A195BXF7 U4UM63 A0A182R8M4 A0A182QH74 A0A1Q3F766 A0A2M4ACF7 Q7Q351 A0A182UE16 A0A2M4AC06 A0A182LPE9 A0A182IWC6 A0A182XC74 A0A182IIF8 A0A182USF3 A0A182K7Q1 A0A195CQJ0 A0A2M4CQL7 W5JT40 A0A182MMK2 A0A182SKE4 A0A182MY14 A0A2M3Z5K9 A0A2M3Z6C7 A0A0T6B415 A0A1Y1KQR1 A0A3F2YWK0 A0A310SS82 A0A182FKQ1 A0A1U7QZJ1 A0A2M4BHX4 A0A2M4BHE3 A0A024R6Z6 Q96MW5 A0A0N0BI72 U4UBE7 K7C7D5 A0A2R8Z9X5 K7CR24
Ontologies
GO
PANTHER
Topology
Subcellular location
Golgi apparatus membrane
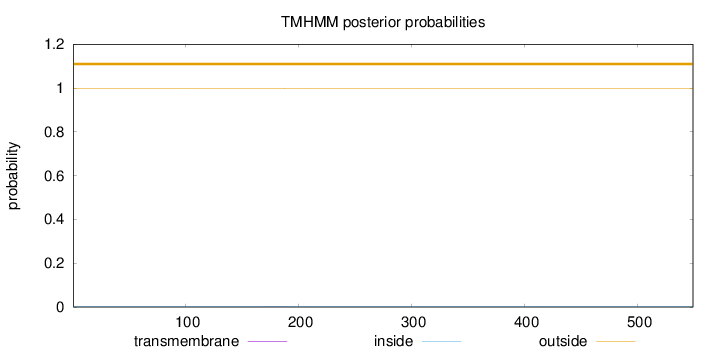
Length:
549
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00941999999999997
Exp number, first 60 AAs:
0.00038
Total prob of N-in:
0.00136
outside
1 - 549
Population Genetic Test Statistics
Pi
143.768034
Theta
148.685411
Tajima's D
-0.966924
CLR
166.330154
CSRT
0.147342632868357
Interpretation
Uncertain
