Gene
KWMTBOMO04505 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005449
Annotation
PREDICTED:_DNA_repair_protein_RAD50_[Bombyx_mori]
Full name
DNA repair protein RAD50
Location in the cell
Nuclear Reliability : 3.84
Sequence
CDS
ATGTTTCAGGCAAAGTACTTTAAATCATTACCCGGGGTTTCAAAAGCCATACTAAATTCTGTCGTATTTTGCCACCAAGAAGATTCAAGCTGGCCACTCGATGAAGGAAAAAAGGTTAAGGAAAGATTTGATGAAATTTTTGATGCTGATAAATACAGTGATTGCTTAGATCGTTTAAAAAAAATAAGAAAGGATTATGCACAGAACTTAAAGTTGCTTGAACAAGAAGTGGCTCATTTAACAGAGAAGAAACAAGATTTAGACAAGAAAAAACTTAATGTAGTTAATACAGAGACAAATATCTCCCAAACTGAAATAAAAGTAACAGAACTAACACAAGAATTAAAACCAGTTAATGAAAAATTAAGTGCTATCAAAACACTCGAGAACAACTTACTGAGTTATGAAACCAAACGAGAAAAAATTAAGATGAGACTAGAGCAAGCTCGAATAGAAGAGGAGAAACTAAAAAACAACATCAAGACTTTGTTTGAAGGAGCACTTGATGAATTGGAACAAAATATAAAAAACTACGCAACTACAGTTAAAGCTAAACAAAAAGAGCTTGAAGATTCATACAAAAAGAACACATCATTCAACAAGGAAGAAGAAAAAATTGCCAATGAGAAGTCTTCAAACGAGATAGAGTACAACAAATTTATATTATTGGAAAGTCAAAATCAAGAGAAGATTGATAAGAGAAATGAACAGATTGTCGAAACTGCAAAAATTGCTGAAATTGAAAACATCACTGAAGTTCAAACAAACGAGGAAGCAGAGAATGCCACAAAAACTATTTTGGAAAAAGTAGAAAGTCTAAAAAGTGACCATAAAGTGCAAAAATCACAAGCTGATGAAGAAGAAAAGAAAGCGCAGGGATTCGTTGATGAATGTCGTGTAGCCTTGTCTGGTCATACACAAAAAATCAGTAGCAAAGAAGGAGAGATAAATAAGAAGAAAAAAGAGATTACTAAAATAGAAAAAGATATAACATCAGCAAATCAATCAAAAGATAAACTAGAATTGATTGAAAAGAAGTTGCTTGCAGTTGAAGAAGAACACAAACAAGCTGAAAACGAATTAAACATTGAAGAATGTCAGCAAGAAATCAACGAAGATGAGAAAGCTGTGGAACAATATGAACAAGAAATGGATGAGCTTAGTCAAAAGATAACCAAGCTTCAAAAGCAAACTGCTAAATTGAAAGAAAAGGAAATGATCGAAGAGAATCTCAAAACTAAAGAAAAACAGTTAATTGTTCTTAAAAACAAACACAAGACGGCCCTGATGGAACTGCTTGGTAATGTACCCGAAAAAGACTTTGCTGTGACAATCAACAAATTTGAGTGTGAAGTGCGAAATGAAGTTGAAACACTGAAGAAAAAATTGAAAGAAAAACAGATAGAGATAACGACTTTGGAAGCGGAGCGAAAACATATTCGTGAATCTTTAAACGAACGACGCAGTGAACTGACTAAAGCCGAAGACAAAATATACAAGGCATGCGGAACCCAGCCCTATGATGTTACGTTATCTAAATATACAACCACAGTAGAAAAACTTCAGGATGAACAAAATGTTTTACAATCTTCAATGTTCATCATAAACAAATATAAAGGGCAACTTAAAGATAACAGTTGCTGTCCATTATGCAATCGCGGCTTTGAAAACGAGTCTGAGGTAACCGACTTAGTAACACAACTAACAACTCAAGTTATGAATGTGCCGGGTAAACTTGAAAAAGTTACTGAAGATCTCCAAAGAGCGTCAGCTAAAAAAGATGAAATACTCAGTATGAAGTCTTTAAATGAGAAAATAACCACTCTTAAAGATACGGAAATCCCCCAGCTGGAGAAACGAATCGTTGATGTTGATACTCAAATTAAGACTCTAAATGAAACTGTGGAAGAAATAACCATGTCTCTGATTGAACCAGAGCAAAAGATGACCACTGCTAAGCAAATACACGGGGATATGCCGATGCTTGACCGCTATATACAAGAAATTAAAACGGTCACCAAAAAGTTTGAGGCAGTAAAAGAACAATGTGGCGATGTTGATTCAGAAATGACGTTGGATGTTGCGACTGCCAAGCAAGTGGATTTTAAGCAAAAAATTAGCACCCTAAGGAATCGCGTAAAAACCACACAGAAGAAACTAAATGCACACAACAAGAAACTGCAAACTTTATATGAAAAAAAAACTAAAGCTAAGGAAGAATTGTTAAATACACAGAAAAAGGTTCAAGACATCGTAAATCTCGAGGAATCTAAAAAGCAACTGCAATCAGATTGTGAGAGACTAGAAACCGAACTAAAAGAACTGAAAGAAGCTATAACCCCACTGGAAGTCGCACTTAAAGAAAAAATCGAGGCTAAGACCGAAGTAGTGAGAAGGAACAGAGATTTGATTGAGGCTGGAAGTGCCTATATTGCAAAAGTCGAAGTAGCTTTCAATAAAGTAAAGTCTCTCGATGCTGAGATCCAACAGCATAAAAAAAAGAATGTACCCCGTGAAATGGAGAAGATACAAGAAGCAAACGATAAATTAATGGAAAAACAAAAACAAATCATGACTGACAGACAAGTGCTCACAAAAAAAATTGATACTCTTAAAGATGAAATTGCAAAGCAAGAGGTTTATAAACGTGATCTAGATGACAACTTGTGTTTGCGTAAAGCACAAACGGACATCACAAATTGGCAAAAACAAGATGATGAATTGAATGAAAAATTGAATGGTATTAACAAAGAAGAATTAAATGAAAAAGAATCATTAATCATTAAGCAAACGAAGTTGTTCCAACAGAAGGCAGAGTGCACTGGAGCATTGAGTGAACTTAAGAAAAGTTTAAAAGGCTTCAGGAATGAACTGGAAAAATCTTTATCTAAGGATATAGAAAAAAAATTTAGAGAAAAAATGTATGAACTACATGTAACTAAAGCTATTGACAAAGATATAAGGGAATATGCTGTTGCTTTAGATAAATGTCTTATGGAGTTTCATAGAGAGAAAATGGAAAATATCAACATGATTATCAGGGAACTTTGGCGAAAGATATACAGAGGTAATGATATTGATTACATTGAAATCAAGACTGAAGGCAACTTAACTGTAGAATCAGAACGACGCAAATATGACTACAGAGTAGTACAATCTAAAAACGGTGTGGAAATCGACATGCGTGGAAGATGTAGTGCTGGTCAAAAAGTACTCGCTTGCCTAATAATAAGGCTAGCACTTGCAGAGACATTCAGTTCCAGGTTTGGAATACTTGCTTTGGATGAACCAACAACAAATTTGGATCAAGAAAACATTCACAGTTTGTGTGCAGCCCTTGGTGAAATAGTCCAAGAGCGAATGATGCAAAAAAACTTTATGTTTATTATTATCACCCATGACAAAGAATTTATAGAGTCACTTGGGAACATTGATAAAGTTACACACTATTATGAGGTGTCTCGAAATGACAATGGCAAATCTAGGGTCAAGAGGATTAGATTTATGTAG
Protein
MFQAKYFKSLPGVSKAILNSVVFCHQEDSSWPLDEGKKVKERFDEIFDADKYSDCLDRLKKIRKDYAQNLKLLEQEVAHLTEKKQDLDKKKLNVVNTETNISQTEIKVTELTQELKPVNEKLSAIKTLENNLLSYETKREKIKMRLEQARIEEEKLKNNIKTLFEGALDELEQNIKNYATTVKAKQKELEDSYKKNTSFNKEEEKIANEKSSNEIEYNKFILLESQNQEKIDKRNEQIVETAKIAEIENITEVQTNEEAENATKTILEKVESLKSDHKVQKSQADEEEKKAQGFVDECRVALSGHTQKISSKEGEINKKKKEITKIEKDITSANQSKDKLELIEKKLLAVEEEHKQAENELNIEECQQEINEDEKAVEQYEQEMDELSQKITKLQKQTAKLKEKEMIEENLKTKEKQLIVLKNKHKTALMELLGNVPEKDFAVTINKFECEVRNEVETLKKKLKEKQIEITTLEAERKHIRESLNERRSELTKAEDKIYKACGTQPYDVTLSKYTTTVEKLQDEQNVLQSSMFIINKYKGQLKDNSCCPLCNRGFENESEVTDLVTQLTTQVMNVPGKLEKVTEDLQRASAKKDEILSMKSLNEKITTLKDTEIPQLEKRIVDVDTQIKTLNETVEEITMSLIEPEQKMTTAKQIHGDMPMLDRYIQEIKTVTKKFEAVKEQCGDVDSEMTLDVATAKQVDFKQKISTLRNRVKTTQKKLNAHNKKLQTLYEKKTKAKEELLNTQKKVQDIVNLEESKKQLQSDCERLETELKELKEAITPLEVALKEKIEAKTEVVRRNRDLIEAGSAYIAKVEVAFNKVKSLDAEIQQHKKKNVPREMEKIQEANDKLMEKQKQIMTDRQVLTKKIDTLKDEIAKQEVYKRDLDDNLCLRKAQTDITNWQKQDDELNEKLNGINKEELNEKESLIIKQTKLFQQKAECTGALSELKKSLKGFRNELEKSLSKDIEKKFREKMYELHVTKAIDKDIREYAVALDKCLMEFHREKMENINMIIRELWRKIYRGNDIDYIEIKTEGNLTVESERRKYDYRVVQSKNGVEIDMRGRCSAGQKVLACLIIRLALAETFSSRFGILALDEPTTNLDQENIHSLCAALGEIVQERMMQKNFMFIIITHDKEFIESLGNIDKVTHYYEVSRNDNGKSRVKRIRFM
Summary
Description
Essential component of the MRN complex, a complex that possesses single-stranded DNA endonuclease and 3' to 5' exonuclease activities, and plays a central role in double-strand break (DSB) repair. The complex participates in processes such as DNA recombination, DNA repair, genome stability, telomere integrity and meiosis. The MRN complex may protect telomeres by facilitating recruitment of HOAP and HP1 at chromosome ends. In the complex, it mediates the ATP-binding and is probably required to bind DNA ends and hold them in close proximity.
Component of the MRN complex, which plays a central role in double-strand break (DSB) repair, DNA recombination, maintenance of telomere integrity and meiosis. The complex possesses single-strand endonuclease activity and double-strand-specific 3'-5' exonuclease activity, which are provided by MRE11. RAD50 may be required to bind DNA ends and hold them in close proximity. This could facilitate searches for short or long regions of sequence homology in the recombining DNA templates, and may also stimulate the activity of DNA ligases and/or restrict the nuclease activity of MRE11 to prevent nucleolytic degradation past a given point. The complex may also be required for DNA damage signaling via activation of the ATM kinase. In telomeres the MRN complex may modulate t-loop formation (By similarity).
Component of the MRN complex, which plays a central role in double-strand break (DSB) repair, DNA recombination, maintenance of telomere integrity and meiosis. The complex possesses single-strand endonuclease activity and double-strand-specific 3'-5' exonuclease activity, which are provided by MRE11. RAD50 may be required to bind DNA ends and hold them in close proximity. This could facilitate searches for short or long regions of sequence homology in the recombining DNA templates, and may also stimulate the activity of DNA ligases and/or restrict the nuclease activity of MRE11 to prevent nucleolytic degradation past a given point. The complex may also be required for DNA damage signaling via activation of the ATM kinase. In telomeres the MRN complex may modulate t-loop formation (By similarity).
Cofactor
Zn(2+)
Subunit
Homodimer. Probable component of the MRN complex with mre11 (By similarity).
Component of the MRN complex composed of two heterodimers RAD50/MRE11 associated with a single NBN. As part of the MRN complex, interacts with MCM8 and MCM9; the interaction recruits the complex to DNA repair sites. Component of the BASC complex, at least composed of BRCA1, MSH2, MSH6, MLH1, ATM, BLM, RAD50, MRE11 and NBN. Found in a complex with TERF2. Interacts with RINT1. Interacts with BRCA1 via its N-terminal domain. Interacts with DCLRE1C/Artemis. Interacts with MRNIP.
Component of the MRN complex composed of two heterodimers RAD50/MRE11 associated with a single NBN. As part of the MRN complex, interacts with MCM8 and MCM9; the interaction recruits the complex to DNA repair sites. Component of the BASC complex, at least composed of BRCA1, MSH2, MSH6, MLH1, ATM, BLM, RAD50, MRE11 and NBN. Found in a complex with TERF2. Interacts with RINT1. Interacts with BRCA1 via its N-terminal domain. Interacts with DCLRE1C/Artemis. Interacts with MRNIP.
Similarity
Belongs to the SMC family. RAD50 subfamily.
Keywords
ATP-binding
Cell cycle
Chromosome
Coiled coil
Complete proteome
DNA damage
DNA repair
Hydrolase
Meiosis
Metal-binding
Nucleotide-binding
Nucleus
Reference proteome
Telomere
Zinc
Acetylation
Alternative splicing
Direct protein sequencing
Phosphoprotein
Feature
chain DNA repair protein RAD50
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
H9J7F7
A0A194PM06
A0A212EI90
A0A2A4J953
A0A194RCL8
A0A067QZL2
+ More
A0A2Z5TRB2 A0A2J7QQY9 A0A2J7QQZ7 A0A2J7QQZ3 A0A1B6F1S1 A0A1B0GH32 A0A0V0G504 A0A2C9JC49 A0A1B6KQF5 A0A1B6C3H1 A0A069DZS5 A0A1B6D4X3 A0A224X5C5 A0A0A1WNN8 A0A0L8GKX7 B4P7X9 A0A1Q3F5U9 A0A182KXV5 A0A0J9U8E7 A0A182YG56 B0WAP9 A0A182QKZ8 A0A0K8UNW1 A0A3B0J5K5 A0A0K8VRS2 E0W0B2 B3NNP5 A0A3B0J5T6 B4MJF7 W8C472 A0A1W4VQG8 W5MWI4 A0A0B4K7L0 D0Z764 A0A1I8NB16 A0A1I8P2K6 A0A3Q0J5N7 A0A3Q0J6C2 V9KA54 Q9W252 A0A3B4DBX2 A0A182G7F0 A0A2I0LTA5 A0A3B4D7T9 B4I847 A0A1U7RTA8 A0A182P3Y5 Q5SV02 A0A3Q3N8A8 P70388 A0A151M734 A0A1A9WRR6 A0A1U7QKB1 A0A084VEJ8 A0A0L0CTW8 F1NEG1 B7NZL1 W5LKZ2 A0A3Q3AN83 A0A087RF50 K9IUU3 A0A182JDN9 A0A0B7AG76 B4QH58 A0A091GX79 F7AR56 A0A3P8TN45 A0A091RC47 A0A3Q1BHM8 A0A091IN49 A0A3Q1BDK4 A0A3B4WWW8 F6RT86 A0A3B5A3G4 A0A3B3QVD8 A0A099YQH8 A0A093C8H2 T1JCH0 A0A093R5B6 A0A0P6J401 A0A3B3QW23 G3V9X6 A0A146P4S6 A0A3B3QSS0 A0A3Q3FJ57 G3WB07 A0A146NW52 A0A1A7WGK8 A0A3P8TJH3 A0A2K6V0L4 A0A2S2QS01 A0A3Q2P9Q3
A0A2Z5TRB2 A0A2J7QQY9 A0A2J7QQZ7 A0A2J7QQZ3 A0A1B6F1S1 A0A1B0GH32 A0A0V0G504 A0A2C9JC49 A0A1B6KQF5 A0A1B6C3H1 A0A069DZS5 A0A1B6D4X3 A0A224X5C5 A0A0A1WNN8 A0A0L8GKX7 B4P7X9 A0A1Q3F5U9 A0A182KXV5 A0A0J9U8E7 A0A182YG56 B0WAP9 A0A182QKZ8 A0A0K8UNW1 A0A3B0J5K5 A0A0K8VRS2 E0W0B2 B3NNP5 A0A3B0J5T6 B4MJF7 W8C472 A0A1W4VQG8 W5MWI4 A0A0B4K7L0 D0Z764 A0A1I8NB16 A0A1I8P2K6 A0A3Q0J5N7 A0A3Q0J6C2 V9KA54 Q9W252 A0A3B4DBX2 A0A182G7F0 A0A2I0LTA5 A0A3B4D7T9 B4I847 A0A1U7RTA8 A0A182P3Y5 Q5SV02 A0A3Q3N8A8 P70388 A0A151M734 A0A1A9WRR6 A0A1U7QKB1 A0A084VEJ8 A0A0L0CTW8 F1NEG1 B7NZL1 W5LKZ2 A0A3Q3AN83 A0A087RF50 K9IUU3 A0A182JDN9 A0A0B7AG76 B4QH58 A0A091GX79 F7AR56 A0A3P8TN45 A0A091RC47 A0A3Q1BHM8 A0A091IN49 A0A3Q1BDK4 A0A3B4WWW8 F6RT86 A0A3B5A3G4 A0A3B3QVD8 A0A099YQH8 A0A093C8H2 T1JCH0 A0A093R5B6 A0A0P6J401 A0A3B3QW23 G3V9X6 A0A146P4S6 A0A3B3QSS0 A0A3Q3FJ57 G3WB07 A0A146NW52 A0A1A7WGK8 A0A3P8TJH3 A0A2K6V0L4 A0A2S2QS01 A0A3Q2P9Q3
EC Number
3.6.-.-
Pubmed
19121390
26354079
22118469
24845553
26760975
15562597
+ More
26334808 25830018 17994087 17550304 20966253 22936249 25244985 20566863 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 24402279 18984573 12537569 15296753 15135728 26483478 23371554 12040188 17525332 19468303 21183079 8910585 16141072 15489334 10377422 12208847 22293439 24438588 26108605 15592404 25329095 19892987 17495919 29240929 15057822 15632090 21709235
26334808 25830018 17994087 17550304 20966253 22936249 25244985 20566863 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 24402279 18984573 12537569 15296753 15135728 26483478 23371554 12040188 17525332 19468303 21183079 8910585 16141072 15489334 10377422 12208847 22293439 24438588 26108605 15592404 25329095 19892987 17495919 29240929 15057822 15632090 21709235
EMBL
BABH01013770
BABH01013771
BABH01013772
KQ459599
KPI94471.1
AGBW02014681
+ More
OWR41199.1 NWSH01002530 PCG68064.1 KQ460397 KPJ15214.1 KK853069 KDR11819.1 FX985773 BBA93660.1 NEVH01012082 PNF31004.1 PNF31005.1 PNF31009.1 GECZ01025672 GECZ01010895 JAS44097.1 JAS58874.1 AJWK01003905 GECL01003006 JAP03118.1 GEBQ01026304 GEBQ01016094 JAT13673.1 JAT23883.1 GEDC01029242 JAS08056.1 GBGD01000130 JAC88759.1 GEDC01016549 JAS20749.1 GFTR01008729 JAW07697.1 GBXI01013628 JAD00664.1 KQ421376 KOF77598.1 CM000158 EDW92134.2 GFDL01012100 JAV22945.1 CM002911 KMY95790.1 DS231874 EDS41690.1 AXCN02001185 GDHF01024116 JAI28198.1 OUUW01000001 SPP75063.1 GDHF01011069 JAI41245.1 DS235858 EEB19068.1 CH954179 EDV55602.2 SPP75062.1 CH963846 EDW72246.1 GAMC01009761 JAB96794.1 AHAT01021397 AE013599 AFH08214.1 BT100311 ACZ52623.1 JW862011 AFO94528.1 BT003555 FJ219324 FJ219329 FJ219330 FJ219331 FJ219332 FJ219333 FJ219334 FJ219335 FJ219336 FJ219337 FJ219338 FJ219339 FJ219340 FJ219341 FJ219342 FJ219343 FJ219344 FJ219345 FJ219346 FJ219347 FJ219348 FJ219349 FJ219350 AY052106 JXUM01046519 KQ561480 KXJ78447.1 AKCR02000104 PKK20650.1 CH480824 EDW56772.1 AL645741 CH466575 EDL33574.1 U66887 AK018001 AK087982 BC058180 AKHW03006437 KYO20250.1 ATLV01012278 KE524778 KFB36392.1 JRES01000027 KNC34829.1 AC159985 DP001045 ACK28136.1 KL226335 KFM12104.1 GABZ01000350 JAA53175.1 HACG01032782 CEK79647.1 CM000362 EDX08204.1 KL513254 KFO87137.1 KK717443 KFQ37393.1 KK500767 KFP10124.1 KL885134 KGL72549.1 KL460192 KFV12235.1 JH432064 KL224978 KFW66199.1 GEBF01006495 JAN97137.1 AC135771 CH473948 EDM04395.1 GCES01147361 JAQ38961.1 AEFK01014865 AEFK01014866 AEFK01014867 AEFK01014868 AEFK01014869 AEFK01014870 GCES01150349 JAQ35973.1 HADW01003296 SBP04696.1 GGMS01010699 MBY79902.1
OWR41199.1 NWSH01002530 PCG68064.1 KQ460397 KPJ15214.1 KK853069 KDR11819.1 FX985773 BBA93660.1 NEVH01012082 PNF31004.1 PNF31005.1 PNF31009.1 GECZ01025672 GECZ01010895 JAS44097.1 JAS58874.1 AJWK01003905 GECL01003006 JAP03118.1 GEBQ01026304 GEBQ01016094 JAT13673.1 JAT23883.1 GEDC01029242 JAS08056.1 GBGD01000130 JAC88759.1 GEDC01016549 JAS20749.1 GFTR01008729 JAW07697.1 GBXI01013628 JAD00664.1 KQ421376 KOF77598.1 CM000158 EDW92134.2 GFDL01012100 JAV22945.1 CM002911 KMY95790.1 DS231874 EDS41690.1 AXCN02001185 GDHF01024116 JAI28198.1 OUUW01000001 SPP75063.1 GDHF01011069 JAI41245.1 DS235858 EEB19068.1 CH954179 EDV55602.2 SPP75062.1 CH963846 EDW72246.1 GAMC01009761 JAB96794.1 AHAT01021397 AE013599 AFH08214.1 BT100311 ACZ52623.1 JW862011 AFO94528.1 BT003555 FJ219324 FJ219329 FJ219330 FJ219331 FJ219332 FJ219333 FJ219334 FJ219335 FJ219336 FJ219337 FJ219338 FJ219339 FJ219340 FJ219341 FJ219342 FJ219343 FJ219344 FJ219345 FJ219346 FJ219347 FJ219348 FJ219349 FJ219350 AY052106 JXUM01046519 KQ561480 KXJ78447.1 AKCR02000104 PKK20650.1 CH480824 EDW56772.1 AL645741 CH466575 EDL33574.1 U66887 AK018001 AK087982 BC058180 AKHW03006437 KYO20250.1 ATLV01012278 KE524778 KFB36392.1 JRES01000027 KNC34829.1 AC159985 DP001045 ACK28136.1 KL226335 KFM12104.1 GABZ01000350 JAA53175.1 HACG01032782 CEK79647.1 CM000362 EDX08204.1 KL513254 KFO87137.1 KK717443 KFQ37393.1 KK500767 KFP10124.1 KL885134 KGL72549.1 KL460192 KFV12235.1 JH432064 KL224978 KFW66199.1 GEBF01006495 JAN97137.1 AC135771 CH473948 EDM04395.1 GCES01147361 JAQ38961.1 AEFK01014865 AEFK01014866 AEFK01014867 AEFK01014868 AEFK01014869 AEFK01014870 GCES01150349 JAQ35973.1 HADW01003296 SBP04696.1 GGMS01010699 MBY79902.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000218220
UP000053240
UP000027135
+ More
UP000235965 UP000092461 UP000076420 UP000053454 UP000002282 UP000075882 UP000076408 UP000002320 UP000075886 UP000268350 UP000009046 UP000008711 UP000007798 UP000192221 UP000018468 UP000000803 UP000095301 UP000095300 UP000079169 UP000261440 UP000069940 UP000249989 UP000053872 UP000001292 UP000189705 UP000075885 UP000000589 UP000261640 UP000050525 UP000091820 UP000189706 UP000030765 UP000037069 UP000000539 UP000018467 UP000264800 UP000053286 UP000075880 UP000000304 UP000002281 UP000265080 UP000257160 UP000053119 UP000261360 UP000002280 UP000261400 UP000261540 UP000053641 UP000054081 UP000002494 UP000007648 UP000233220 UP000265000
UP000235965 UP000092461 UP000076420 UP000053454 UP000002282 UP000075882 UP000076408 UP000002320 UP000075886 UP000268350 UP000009046 UP000008711 UP000007798 UP000192221 UP000018468 UP000000803 UP000095301 UP000095300 UP000079169 UP000261440 UP000069940 UP000249989 UP000053872 UP000001292 UP000189705 UP000075885 UP000000589 UP000261640 UP000050525 UP000091820 UP000189706 UP000030765 UP000037069 UP000000539 UP000018467 UP000264800 UP000053286 UP000075880 UP000000304 UP000002281 UP000265080 UP000257160 UP000053119 UP000261360 UP000002280 UP000261400 UP000261540 UP000053641 UP000054081 UP000002494 UP000007648 UP000233220 UP000265000
Interpro
ProteinModelPortal
H9J7F7
A0A194PM06
A0A212EI90
A0A2A4J953
A0A194RCL8
A0A067QZL2
+ More
A0A2Z5TRB2 A0A2J7QQY9 A0A2J7QQZ7 A0A2J7QQZ3 A0A1B6F1S1 A0A1B0GH32 A0A0V0G504 A0A2C9JC49 A0A1B6KQF5 A0A1B6C3H1 A0A069DZS5 A0A1B6D4X3 A0A224X5C5 A0A0A1WNN8 A0A0L8GKX7 B4P7X9 A0A1Q3F5U9 A0A182KXV5 A0A0J9U8E7 A0A182YG56 B0WAP9 A0A182QKZ8 A0A0K8UNW1 A0A3B0J5K5 A0A0K8VRS2 E0W0B2 B3NNP5 A0A3B0J5T6 B4MJF7 W8C472 A0A1W4VQG8 W5MWI4 A0A0B4K7L0 D0Z764 A0A1I8NB16 A0A1I8P2K6 A0A3Q0J5N7 A0A3Q0J6C2 V9KA54 Q9W252 A0A3B4DBX2 A0A182G7F0 A0A2I0LTA5 A0A3B4D7T9 B4I847 A0A1U7RTA8 A0A182P3Y5 Q5SV02 A0A3Q3N8A8 P70388 A0A151M734 A0A1A9WRR6 A0A1U7QKB1 A0A084VEJ8 A0A0L0CTW8 F1NEG1 B7NZL1 W5LKZ2 A0A3Q3AN83 A0A087RF50 K9IUU3 A0A182JDN9 A0A0B7AG76 B4QH58 A0A091GX79 F7AR56 A0A3P8TN45 A0A091RC47 A0A3Q1BHM8 A0A091IN49 A0A3Q1BDK4 A0A3B4WWW8 F6RT86 A0A3B5A3G4 A0A3B3QVD8 A0A099YQH8 A0A093C8H2 T1JCH0 A0A093R5B6 A0A0P6J401 A0A3B3QW23 G3V9X6 A0A146P4S6 A0A3B3QSS0 A0A3Q3FJ57 G3WB07 A0A146NW52 A0A1A7WGK8 A0A3P8TJH3 A0A2K6V0L4 A0A2S2QS01 A0A3Q2P9Q3
A0A2Z5TRB2 A0A2J7QQY9 A0A2J7QQZ7 A0A2J7QQZ3 A0A1B6F1S1 A0A1B0GH32 A0A0V0G504 A0A2C9JC49 A0A1B6KQF5 A0A1B6C3H1 A0A069DZS5 A0A1B6D4X3 A0A224X5C5 A0A0A1WNN8 A0A0L8GKX7 B4P7X9 A0A1Q3F5U9 A0A182KXV5 A0A0J9U8E7 A0A182YG56 B0WAP9 A0A182QKZ8 A0A0K8UNW1 A0A3B0J5K5 A0A0K8VRS2 E0W0B2 B3NNP5 A0A3B0J5T6 B4MJF7 W8C472 A0A1W4VQG8 W5MWI4 A0A0B4K7L0 D0Z764 A0A1I8NB16 A0A1I8P2K6 A0A3Q0J5N7 A0A3Q0J6C2 V9KA54 Q9W252 A0A3B4DBX2 A0A182G7F0 A0A2I0LTA5 A0A3B4D7T9 B4I847 A0A1U7RTA8 A0A182P3Y5 Q5SV02 A0A3Q3N8A8 P70388 A0A151M734 A0A1A9WRR6 A0A1U7QKB1 A0A084VEJ8 A0A0L0CTW8 F1NEG1 B7NZL1 W5LKZ2 A0A3Q3AN83 A0A087RF50 K9IUU3 A0A182JDN9 A0A0B7AG76 B4QH58 A0A091GX79 F7AR56 A0A3P8TN45 A0A091RC47 A0A3Q1BHM8 A0A091IN49 A0A3Q1BDK4 A0A3B4WWW8 F6RT86 A0A3B5A3G4 A0A3B3QVD8 A0A099YQH8 A0A093C8H2 T1JCH0 A0A093R5B6 A0A0P6J401 A0A3B3QW23 G3V9X6 A0A146P4S6 A0A3B3QSS0 A0A3Q3FJ57 G3WB07 A0A146NW52 A0A1A7WGK8 A0A3P8TJH3 A0A2K6V0L4 A0A2S2QS01 A0A3Q2P9Q3
PDB
5DA9
E-value=8.55226e-40,
Score=416
Ontologies
GO
GO:0046872
GO:0030870
GO:0006281
GO:0000723
GO:0016887
GO:0005524
GO:0006302
GO:0016233
GO:0000722
GO:0008104
GO:0000793
GO:0032508
GO:0070192
GO:0000784
GO:0007004
GO:0000794
GO:0035861
GO:0051880
GO:0090305
GO:0043047
GO:0000790
GO:0004017
GO:0003691
GO:0003690
GO:0051321
GO:0000781
GO:0051276
GO:0005634
GO:0005654
GO:0032206
GO:0045120
GO:0007346
GO:0033674
GO:0016234
GO:0003677
GO:0000019
GO:0031954
GO:0006974
GO:1904354
GO:0030674
GO:0031860
GO:0046597
GO:0051291
GO:0048471
GO:0006310
GO:0006955
GO:0005136
GO:0005576
GO:0008083
GO:0006418
GO:0009058
GO:0016742
GO:0003824
Topology
Subcellular location
Nucleus
Chromosome
Telomere
Chromosome
Telomere
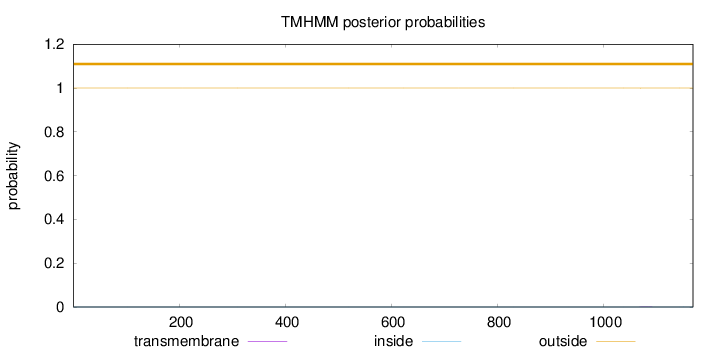
Length:
1169
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00717999999999999
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00001
outside
1 - 1169
Population Genetic Test Statistics
Pi
187.04012
Theta
183.74862
Tajima's D
0.025857
CLR
0.277492
CSRT
0.370381480925954
Interpretation
Uncertain
