Gene
KWMTBOMO04503
Pre Gene Modal
BGIBMGA005362
Annotation
PREDICTED:_transmembrane_protein_47_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.897
Sequence
CDS
ATGATCGAGACTGTTACCATAACAAGGCCATTAAAGGTAATCGCGCTAATATGTGGCTTGCTGGTTGTAATCCTGATGGTGCTTGGACTGGCATCAGCGGACTGGCTAATGGCGGCCGGCTGGCGTCAGGGGCTTTTTATGCACTGCATCGACCCAATCGCACCAACACCCTTGCCCTTCGACATTATAGCTCAGCCGGGATGCTACGCCGCGAGACCAGCCCCATACATCAAGGCCGCGGCTGGTTTGTGCGTGGCGACGCTAGCGGCGGATGTGTGCGGGGCTCTGTTGACCGGGCTCGGACTGCGCTCCACTGATCACCGCACTAAGTTTCGGTACTACCGTTTCGCAGTGCTCGCTATGGCCCTTGCACTGATGTGCATTTTGATAGCATTAGTCATCTACCCGGTTTGCTTTGCGGCCGAATTAAATCTGGGTAAGCGAATTATGTTTTTCTTTTTAATTTCTATCAATTGTTATATTCTAAAGATATGGTATGGTATTTTTAATAAAGGTATGGTATTTTCTTTTCGTATTTTTATATAA
Protein
MIETVTITRPLKVIALICGLLVVILMVLGLASADWLMAAGWRQGLFMHCIDPIAPTPLPFDIIAQPGCYAARPAPYIKAAAGLCVATLAADVCGALLTGLGLRSTDHRTKFRYYRFAVLAMALALMCILIALVIYPVCFAAELNLGKRIMFFFLISINCYILKIWYGIFNKGMVFSFRIFI
Summary
Uniprot
A0A194RC82
A0A0A9YAK7
A0A2H8TMB3
J9JQQ8
A0A158NB47
A0A0C9RY15
+ More
E0VCG3 A0A139WBQ2 E9J006 A0A026W1Y3 A0A1L8D8N2 A0A1Y1KNE9 A0A1B6GWQ5 A0A1L8D8X7 A0A1B6IVK7 A0A2S2QWQ8 K7IVH9 A0A0P4WJB9 A0A1B6MJZ0 A0A0A1WQN3 A0A0K8UB50 A0A034VKH6 W8BS79 A0A336MHU7 A0A1B6M2K5 A0A0Q9WYR2 A0A224XT87 A0A069DX37 A0A034VIU2 A0A1Q3FBY0 B0XA51 W8BLR1 B4LXC2 B4N9Q4 A0A0M4EPG3 A0A0P8XZR6 A0A1B6CXG9 A0A1I8MWC8 A0A1I8Q7M0 A0A0R1E9D0 U5EY49 B4JIE9 B4KAC0 B4IQP5 A0A1Q3FVL6 A0A1W4V8P2 A0A0Q9WTM5 A0A1B6DFK5 B4HET1 A0A3B0KH50 B3LZR1 B4PME8 Q17L63 A0A1A9XG99 A0A1A9ZHM5 D3TS26 B4R0Q4 Q9VCV1 A0A1I8Q7J7 A0A182H7W2 A0A1I8MWC9 A0A023ELH6 A0A0R3NJQ0 A0A1W4VKE7 B3P8K3
E0VCG3 A0A139WBQ2 E9J006 A0A026W1Y3 A0A1L8D8N2 A0A1Y1KNE9 A0A1B6GWQ5 A0A1L8D8X7 A0A1B6IVK7 A0A2S2QWQ8 K7IVH9 A0A0P4WJB9 A0A1B6MJZ0 A0A0A1WQN3 A0A0K8UB50 A0A034VKH6 W8BS79 A0A336MHU7 A0A1B6M2K5 A0A0Q9WYR2 A0A224XT87 A0A069DX37 A0A034VIU2 A0A1Q3FBY0 B0XA51 W8BLR1 B4LXC2 B4N9Q4 A0A0M4EPG3 A0A0P8XZR6 A0A1B6CXG9 A0A1I8MWC8 A0A1I8Q7M0 A0A0R1E9D0 U5EY49 B4JIE9 B4KAC0 B4IQP5 A0A1Q3FVL6 A0A1W4V8P2 A0A0Q9WTM5 A0A1B6DFK5 B4HET1 A0A3B0KH50 B3LZR1 B4PME8 Q17L63 A0A1A9XG99 A0A1A9ZHM5 D3TS26 B4R0Q4 Q9VCV1 A0A1I8Q7J7 A0A182H7W2 A0A1I8MWC9 A0A023ELH6 A0A0R3NJQ0 A0A1W4VKE7 B3P8K3
Pubmed
26354079
25401762
21347285
20566863
18362917
19820115
+ More
21282665 24508170 28004739 20075255 25830018 25348373 24495485 17994087 26334808 18057021 25315136 17550304 17510324 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 24945155 15632085
21282665 24508170 28004739 20075255 25830018 25348373 24495485 17994087 26334808 18057021 25315136 17550304 17510324 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 24945155 15632085
EMBL
KQ460397
KPJ15212.1
GBHO01013512
JAG30092.1
GFXV01002997
MBW14802.1
+ More
ABLF02038155 ABLF02038161 ADTU01010876 GBYB01012781 GBYB01012784 JAG82548.1 JAG82551.1 DS235053 EEB11069.1 KQ971372 KYB25333.1 GL767291 EFZ13877.1 KK107485 EZA50080.1 GFDF01011248 JAV02836.1 GEZM01081259 JAV61730.1 GECZ01018454 GECZ01002895 JAS51315.1 JAS66874.1 GFDF01011171 JAV02913.1 GECU01016758 JAS90948.1 GGMS01012931 MBY82134.1 GDRN01068559 JAI64179.1 GEBQ01003756 JAT36221.1 GBXI01015462 GBXI01013331 GBXI01008635 JAC98829.1 JAD00961.1 JAD05657.1 GDHF01028417 GDHF01022591 GDHF01006351 JAI23897.1 JAI29723.1 JAI45963.1 GAKP01016901 GAKP01016899 GAKP01016898 GAKP01016897 JAC42053.1 GAMC01006802 GAMC01006801 JAB99753.1 UFQS01001232 UFQT01001232 SSX09872.1 SSX29595.1 GEBQ01009798 JAT30179.1 CH933806 KRG01104.1 GFTR01003388 JAW13038.1 GBGD01002995 JAC85894.1 GAKP01016900 JAC42052.1 GFDL01009961 JAV25084.1 DS232564 EDS43516.1 GAMC01006803 JAB99752.1 CH940650 EDW67800.1 CH964232 EDW80619.2 KRF99312.1 CP012526 ALC45997.1 CH902617 KPU80047.1 GEDC01019283 JAS18015.1 CM000160 KRK04096.1 KRK04098.1 GANO01000602 JAB59269.1 CH916369 EDV93030.1 EDW14607.2 CH689249 EDW45286.1 GFDL01003529 JAV31516.1 KRF83476.1 KRF83477.1 GEDC01012817 JAS24481.1 CH480815 EDW43242.1 OUUW01000007 SPP82998.1 EDV43055.2 EDW98053.2 CH477218 EAT47432.1 CCAG010010579 EZ424228 ADD20504.1 CM000364 EDX13977.1 AE014297 BT011196 AAF56054.1 AAR88558.1 JXUM01028359 JXUM01028360 JXUM01028361 JXUM01028362 KQ560819 KXJ80763.1 GAPW01004499 JAC09099.1 CM000070 KRS99886.1 CH954182 EDV54098.1
ABLF02038155 ABLF02038161 ADTU01010876 GBYB01012781 GBYB01012784 JAG82548.1 JAG82551.1 DS235053 EEB11069.1 KQ971372 KYB25333.1 GL767291 EFZ13877.1 KK107485 EZA50080.1 GFDF01011248 JAV02836.1 GEZM01081259 JAV61730.1 GECZ01018454 GECZ01002895 JAS51315.1 JAS66874.1 GFDF01011171 JAV02913.1 GECU01016758 JAS90948.1 GGMS01012931 MBY82134.1 GDRN01068559 JAI64179.1 GEBQ01003756 JAT36221.1 GBXI01015462 GBXI01013331 GBXI01008635 JAC98829.1 JAD00961.1 JAD05657.1 GDHF01028417 GDHF01022591 GDHF01006351 JAI23897.1 JAI29723.1 JAI45963.1 GAKP01016901 GAKP01016899 GAKP01016898 GAKP01016897 JAC42053.1 GAMC01006802 GAMC01006801 JAB99753.1 UFQS01001232 UFQT01001232 SSX09872.1 SSX29595.1 GEBQ01009798 JAT30179.1 CH933806 KRG01104.1 GFTR01003388 JAW13038.1 GBGD01002995 JAC85894.1 GAKP01016900 JAC42052.1 GFDL01009961 JAV25084.1 DS232564 EDS43516.1 GAMC01006803 JAB99752.1 CH940650 EDW67800.1 CH964232 EDW80619.2 KRF99312.1 CP012526 ALC45997.1 CH902617 KPU80047.1 GEDC01019283 JAS18015.1 CM000160 KRK04096.1 KRK04098.1 GANO01000602 JAB59269.1 CH916369 EDV93030.1 EDW14607.2 CH689249 EDW45286.1 GFDL01003529 JAV31516.1 KRF83476.1 KRF83477.1 GEDC01012817 JAS24481.1 CH480815 EDW43242.1 OUUW01000007 SPP82998.1 EDV43055.2 EDW98053.2 CH477218 EAT47432.1 CCAG010010579 EZ424228 ADD20504.1 CM000364 EDX13977.1 AE014297 BT011196 AAF56054.1 AAR88558.1 JXUM01028359 JXUM01028360 JXUM01028361 JXUM01028362 KQ560819 KXJ80763.1 GAPW01004499 JAC09099.1 CM000070 KRS99886.1 CH954182 EDV54098.1
Proteomes
UP000053240
UP000007819
UP000005205
UP000009046
UP000007266
UP000053097
+ More
UP000002358 UP000009192 UP000002320 UP000008792 UP000007798 UP000092553 UP000007801 UP000095301 UP000095300 UP000002282 UP000001070 UP000001292 UP000192221 UP000268350 UP000008820 UP000092443 UP000092445 UP000092444 UP000000304 UP000000803 UP000069940 UP000249989 UP000001819 UP000008711
UP000002358 UP000009192 UP000002320 UP000008792 UP000007798 UP000092553 UP000007801 UP000095301 UP000095300 UP000002282 UP000001070 UP000001292 UP000192221 UP000268350 UP000008820 UP000092443 UP000092445 UP000092444 UP000000304 UP000000803 UP000069940 UP000249989 UP000001819 UP000008711
PRIDE
Pfam
PF00822 PMP22_Claudin
ProteinModelPortal
A0A194RC82
A0A0A9YAK7
A0A2H8TMB3
J9JQQ8
A0A158NB47
A0A0C9RY15
+ More
E0VCG3 A0A139WBQ2 E9J006 A0A026W1Y3 A0A1L8D8N2 A0A1Y1KNE9 A0A1B6GWQ5 A0A1L8D8X7 A0A1B6IVK7 A0A2S2QWQ8 K7IVH9 A0A0P4WJB9 A0A1B6MJZ0 A0A0A1WQN3 A0A0K8UB50 A0A034VKH6 W8BS79 A0A336MHU7 A0A1B6M2K5 A0A0Q9WYR2 A0A224XT87 A0A069DX37 A0A034VIU2 A0A1Q3FBY0 B0XA51 W8BLR1 B4LXC2 B4N9Q4 A0A0M4EPG3 A0A0P8XZR6 A0A1B6CXG9 A0A1I8MWC8 A0A1I8Q7M0 A0A0R1E9D0 U5EY49 B4JIE9 B4KAC0 B4IQP5 A0A1Q3FVL6 A0A1W4V8P2 A0A0Q9WTM5 A0A1B6DFK5 B4HET1 A0A3B0KH50 B3LZR1 B4PME8 Q17L63 A0A1A9XG99 A0A1A9ZHM5 D3TS26 B4R0Q4 Q9VCV1 A0A1I8Q7J7 A0A182H7W2 A0A1I8MWC9 A0A023ELH6 A0A0R3NJQ0 A0A1W4VKE7 B3P8K3
E0VCG3 A0A139WBQ2 E9J006 A0A026W1Y3 A0A1L8D8N2 A0A1Y1KNE9 A0A1B6GWQ5 A0A1L8D8X7 A0A1B6IVK7 A0A2S2QWQ8 K7IVH9 A0A0P4WJB9 A0A1B6MJZ0 A0A0A1WQN3 A0A0K8UB50 A0A034VKH6 W8BS79 A0A336MHU7 A0A1B6M2K5 A0A0Q9WYR2 A0A224XT87 A0A069DX37 A0A034VIU2 A0A1Q3FBY0 B0XA51 W8BLR1 B4LXC2 B4N9Q4 A0A0M4EPG3 A0A0P8XZR6 A0A1B6CXG9 A0A1I8MWC8 A0A1I8Q7M0 A0A0R1E9D0 U5EY49 B4JIE9 B4KAC0 B4IQP5 A0A1Q3FVL6 A0A1W4V8P2 A0A0Q9WTM5 A0A1B6DFK5 B4HET1 A0A3B0KH50 B3LZR1 B4PME8 Q17L63 A0A1A9XG99 A0A1A9ZHM5 D3TS26 B4R0Q4 Q9VCV1 A0A1I8Q7J7 A0A182H7W2 A0A1I8MWC9 A0A023ELH6 A0A0R3NJQ0 A0A1W4VKE7 B3P8K3
Ontologies
PANTHER
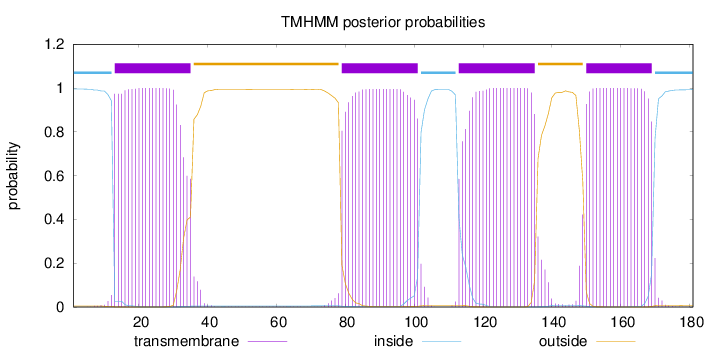
Topology
Length:
181
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
88.15767
Exp number, first 60 AAs:
22.01721
Total prob of N-in:
0.99531
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 78
TMhelix
79 - 101
inside
102 - 112
TMhelix
113 - 135
outside
136 - 149
TMhelix
150 - 169
inside
170 - 181
Population Genetic Test Statistics
Pi
177.294294
Theta
169.769192
Tajima's D
0.070298
CLR
0.514069
CSRT
0.386580670966452
Interpretation
Uncertain
