Gene
KWMTBOMO04501
Pre Gene Modal
BGIBMGA005447
Annotation
PREDICTED:_gamma-glutamyl_transpeptidase_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.601 PlasmaMembrane Reliability : 1.115
Sequence
CDS
ATGCTACGCGGATGGATCGGCGTCGAGACCGTGGCTCCCGTAGGCTCCGACCTATACTGGCATCGCATCGTGGAAACCTTCAAATACGCGTACGCGAAGCGAACAGGCCTCGGGGACCCTTCACGTCACAACCTTAACCAAAGTATTCTTGAGTTAGAGCAAAACCTTGCAAATGCTGAGTGGGCTGCCTCGTATAGAGGGCGCGTGGACGACACCCGCACATACAATGATTGGAAACACTATGAGGCTGTGTTCGAGGGAGCAGACGATCACGGAACAGCGCACGTCATCGTTGTGGCGCCCGACGGAAGCGTGGTCTCTGCAACCAGCACTATCAACTTTATATGGGGCTCGCAAAGAAGGTCTCAAAGTTTAGGCTTCATACTCAACAATGAGATGGATGATTTCGCCATTCCGAATCGAGACTCGTCATATGGACTTCCCCCTTCGCCGGCCAACATGCTAGCTCCTGGACTACAGCCTCTCAGTTCAATGGTGCCCAGTGTTGTTGTGTCAAAGAGCGGAGCTGCAGATTTAGTTTTGGGTGCAGCCGGTGGCACTAAAATTACAACTCAAGTCGCATTGATGGCGACACACATCATAGTCGAGGGCCACGACCTCCCGGATGTGATTCGGCGACCTCGATTACATCATCAACTGTTGCCGATGGAAATTCAACACGAAGCTAATTTCGATTCAGACGTAATAGCATCATTGCGTAGCAAAGGACATGCAACAACTGAACTGGGACCCACGGCGGGTTTCGCTGCAATGGTGGCCATACACCGTGATCGCTCGGGATATTTGGTCCCGGCCACAGATCACAGAAGAATCGTTCGCCATATTTAG
Protein
MLRGWIGVETVAPVGSDLYWHRIVETFKYAYAKRTGLGDPSRHNLNQSILELEQNLANAEWAASYRGRVDDTRTYNDWKHYEAVFEGADDHGTAHVIVVAPDGSVVSATSTINFIWGSQRRSQSLGFILNNEMDDFAIPNRDSSYGLPPSPANMLAPGLQPLSSMVPSVVVSKSGAADLVLGAAGGTKITTQVALMATHIIVEGHDLPDVIRRPRLHHQLLPMEIQHEANFDSDVIASLRSKGHATTELGPTAGFAAMVAIHRDRSGYLVPATDHRRIVRHI
Summary
Uniprot
H9J7F5
F8V3L0
A0A2A4J9C5
A0A2H1WY09
A0A212EI86
A0A194RCL1
+ More
A0A194PMC0 A0A3S2M5M0 A0A0L7LCA7 A0A1L8E275 A0A1L8E2D3 A0A1B0D2V9 B0W7T9 A0A2M3Z3J8 A0A1Q3FF98 A0A1Q3FF50 A0A2M3Z3Q9 A0A182GK30 A0A2M4AH88 A0A2M4AH32 A0A023EW61 A0A182GCH1 A0A2M4BIR4 A0A2M4BII3 W5JRT3 B0W7U0 A0A2M4BIN3 A0A182SZG7 A0A182LXZ4 A0A1Q3F715 A0A1Q3F6X3 A0A182VUE3 A0A336N0K6 A0A182HCT7 A0A182KJU5 Q17DD8 A0A182GK31 A0A1S4F713 A0A182F1D3 A0A336LD73 A0A182LMU2 V9IK84 A0A182KEL7 A0A182U044 A0A182V7Y3 A0A1S4HED3 A0A182RIR1 A0A084W5I9 A0A182Y0C0 E2BSL0 U5EWG4 A0A182XIW5 Q7Q0N5 A0A1S4H3F0 A0A0K8TPF4 A0A182I7B4 A0A182XNQ6 A0A182I0S9 K7J8Z0 A0A182N9R3 A0A182MN44 A0A182MZ66 A0A195EFZ2 A0A088A315 A0A182Q6K4 A0A084W7D9 A0A336KBV3 A0A182T2W5 A0A182QND5 A0A232EJ49 A0A182JQB1 A0A182FMX1 W5JA71 E2AC21 A0A182J7S8 A0A182V766 A0A084W7E0 A0A182KEE7 A0A2M4AGU2 A0A151I564 A0A2M4AH12 A0A0J7KHC8 A0A195CKM7 Q7PWM4 A0A2M3Z6V2 A0A182N8S8 Q16NY3 A0A182VSW2 A0A151WJM9 A0A182GCH0 A0A195F6Q3 A0A1S4H0D0 Q16RJ3 W5JRA5 A0A2M3Z2Q5 A0A2M3Z2U7 A0A182U7D5 A0A2M4BIX4 A0A2M4BIL8 A0A2M4BIM3
A0A194PMC0 A0A3S2M5M0 A0A0L7LCA7 A0A1L8E275 A0A1L8E2D3 A0A1B0D2V9 B0W7T9 A0A2M3Z3J8 A0A1Q3FF98 A0A1Q3FF50 A0A2M3Z3Q9 A0A182GK30 A0A2M4AH88 A0A2M4AH32 A0A023EW61 A0A182GCH1 A0A2M4BIR4 A0A2M4BII3 W5JRT3 B0W7U0 A0A2M4BIN3 A0A182SZG7 A0A182LXZ4 A0A1Q3F715 A0A1Q3F6X3 A0A182VUE3 A0A336N0K6 A0A182HCT7 A0A182KJU5 Q17DD8 A0A182GK31 A0A1S4F713 A0A182F1D3 A0A336LD73 A0A182LMU2 V9IK84 A0A182KEL7 A0A182U044 A0A182V7Y3 A0A1S4HED3 A0A182RIR1 A0A084W5I9 A0A182Y0C0 E2BSL0 U5EWG4 A0A182XIW5 Q7Q0N5 A0A1S4H3F0 A0A0K8TPF4 A0A182I7B4 A0A182XNQ6 A0A182I0S9 K7J8Z0 A0A182N9R3 A0A182MN44 A0A182MZ66 A0A195EFZ2 A0A088A315 A0A182Q6K4 A0A084W7D9 A0A336KBV3 A0A182T2W5 A0A182QND5 A0A232EJ49 A0A182JQB1 A0A182FMX1 W5JA71 E2AC21 A0A182J7S8 A0A182V766 A0A084W7E0 A0A182KEE7 A0A2M4AGU2 A0A151I564 A0A2M4AH12 A0A0J7KHC8 A0A195CKM7 Q7PWM4 A0A2M3Z6V2 A0A182N8S8 Q16NY3 A0A182VSW2 A0A151WJM9 A0A182GCH0 A0A195F6Q3 A0A1S4H0D0 Q16RJ3 W5JRA5 A0A2M3Z2Q5 A0A2M3Z2U7 A0A182U7D5 A0A2M4BIX4 A0A2M4BIL8 A0A2M4BIM3
Pubmed
EMBL
BABH01013764
JN021673
AEJ33054.1
NWSH01002530
PCG68060.1
ODYU01011926
+ More
SOQ57949.1 AGBW02014681 OWR41202.1 KQ460397 KPJ15209.1 KQ459599 KPI94467.1 RSAL01000032 RVE51524.1 JTDY01001700 KOB73123.1 GFDF01001277 JAV12807.1 GFDF01001278 JAV12806.1 AJVK01002880 DS231855 EDS38233.1 GGFM01002348 MBW23099.1 GFDL01008816 JAV26229.1 GFDL01008903 JAV26142.1 GGFM01002403 MBW23154.1 JXUM01013073 KQ560368 KXJ82764.1 GGFK01006835 MBW40156.1 GGFK01006778 MBW40099.1 GAPW01000989 JAC12609.1 JXUM01054221 KQ561811 KXJ77452.1 GGFJ01003700 MBW52841.1 GGFJ01003701 MBW52842.1 ADMH02000654 ETN65434.1 EDS38234.1 GGFJ01003702 MBW52843.1 AXCM01003842 GFDL01011732 JAV23313.1 GFDL01011735 JAV23310.1 UFQT01003036 SSX34473.1 JXUM01128233 KQ567291 KXJ69487.1 CH477296 EAT44399.1 JXUM01013074 KXJ82765.1 UFQS01002842 UFQT01002842 SSX14727.1 SSX34122.1 JR048636 AEY60714.1 AAAB01008984 ATLV01020656 KE525304 KFB45483.1 GL450240 EFN81343.1 GANO01000588 JAB59283.1 AAAB01008980 EAA13957.4 GDAI01001828 JAI15775.1 APCN01003376 APCN01003377 APCN01005255 APCN01005256 APCN01005257 AXCM01000683 KQ978957 KYN27158.1 AXCN02002068 ATLV01021228 KE525315 KFB46133.1 UFQS01000134 UFQT01000134 SSX00273.1 SSX20653.1 AXCN02001863 NNAY01004090 OXU18386.1 ADMH02002068 ETN59664.1 GL438389 EFN69039.1 KFB46134.1 GGFK01006611 MBW39932.1 KQ976456 KYM84867.1 GGFK01006750 MBW40071.1 LBMM01007319 KMQ89853.1 KQ977622 KYN01276.1 EAA14856.5 GGFM01003505 MBW24256.1 CH477802 EAT36069.1 KQ983039 KYQ48069.1 KXJ77451.1 KQ981763 KYN36061.1 CH477708 EAT37027.1 ETN65435.1 GGFM01002034 MBW22785.1 GGFM01002027 MBW22778.1 GGFJ01003760 MBW52901.1 GGFJ01003759 MBW52900.1 GGFJ01003670 MBW52811.1
SOQ57949.1 AGBW02014681 OWR41202.1 KQ460397 KPJ15209.1 KQ459599 KPI94467.1 RSAL01000032 RVE51524.1 JTDY01001700 KOB73123.1 GFDF01001277 JAV12807.1 GFDF01001278 JAV12806.1 AJVK01002880 DS231855 EDS38233.1 GGFM01002348 MBW23099.1 GFDL01008816 JAV26229.1 GFDL01008903 JAV26142.1 GGFM01002403 MBW23154.1 JXUM01013073 KQ560368 KXJ82764.1 GGFK01006835 MBW40156.1 GGFK01006778 MBW40099.1 GAPW01000989 JAC12609.1 JXUM01054221 KQ561811 KXJ77452.1 GGFJ01003700 MBW52841.1 GGFJ01003701 MBW52842.1 ADMH02000654 ETN65434.1 EDS38234.1 GGFJ01003702 MBW52843.1 AXCM01003842 GFDL01011732 JAV23313.1 GFDL01011735 JAV23310.1 UFQT01003036 SSX34473.1 JXUM01128233 KQ567291 KXJ69487.1 CH477296 EAT44399.1 JXUM01013074 KXJ82765.1 UFQS01002842 UFQT01002842 SSX14727.1 SSX34122.1 JR048636 AEY60714.1 AAAB01008984 ATLV01020656 KE525304 KFB45483.1 GL450240 EFN81343.1 GANO01000588 JAB59283.1 AAAB01008980 EAA13957.4 GDAI01001828 JAI15775.1 APCN01003376 APCN01003377 APCN01005255 APCN01005256 APCN01005257 AXCM01000683 KQ978957 KYN27158.1 AXCN02002068 ATLV01021228 KE525315 KFB46133.1 UFQS01000134 UFQT01000134 SSX00273.1 SSX20653.1 AXCN02001863 NNAY01004090 OXU18386.1 ADMH02002068 ETN59664.1 GL438389 EFN69039.1 KFB46134.1 GGFK01006611 MBW39932.1 KQ976456 KYM84867.1 GGFK01006750 MBW40071.1 LBMM01007319 KMQ89853.1 KQ977622 KYN01276.1 EAA14856.5 GGFM01003505 MBW24256.1 CH477802 EAT36069.1 KQ983039 KYQ48069.1 KXJ77451.1 KQ981763 KYN36061.1 CH477708 EAT37027.1 ETN65435.1 GGFM01002034 MBW22785.1 GGFM01002027 MBW22778.1 GGFJ01003760 MBW52901.1 GGFJ01003759 MBW52900.1 GGFJ01003670 MBW52811.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000283053
+ More
UP000037510 UP000092462 UP000002320 UP000069940 UP000249989 UP000000673 UP000075901 UP000075883 UP000075920 UP000075882 UP000008820 UP000069272 UP000075881 UP000075902 UP000075903 UP000075900 UP000030765 UP000076408 UP000008237 UP000076407 UP000007062 UP000075840 UP000002358 UP000075884 UP000078492 UP000005203 UP000075886 UP000215335 UP000000311 UP000075880 UP000078540 UP000036403 UP000078542 UP000075809 UP000078541
UP000037510 UP000092462 UP000002320 UP000069940 UP000249989 UP000000673 UP000075901 UP000075883 UP000075920 UP000075882 UP000008820 UP000069272 UP000075881 UP000075902 UP000075903 UP000075900 UP000030765 UP000076408 UP000008237 UP000076407 UP000007062 UP000075840 UP000002358 UP000075884 UP000078492 UP000005203 UP000075886 UP000215335 UP000000311 UP000075880 UP000078540 UP000036403 UP000078542 UP000075809 UP000078541
Interpro
Gene 3D
ProteinModelPortal
H9J7F5
F8V3L0
A0A2A4J9C5
A0A2H1WY09
A0A212EI86
A0A194RCL1
+ More
A0A194PMC0 A0A3S2M5M0 A0A0L7LCA7 A0A1L8E275 A0A1L8E2D3 A0A1B0D2V9 B0W7T9 A0A2M3Z3J8 A0A1Q3FF98 A0A1Q3FF50 A0A2M3Z3Q9 A0A182GK30 A0A2M4AH88 A0A2M4AH32 A0A023EW61 A0A182GCH1 A0A2M4BIR4 A0A2M4BII3 W5JRT3 B0W7U0 A0A2M4BIN3 A0A182SZG7 A0A182LXZ4 A0A1Q3F715 A0A1Q3F6X3 A0A182VUE3 A0A336N0K6 A0A182HCT7 A0A182KJU5 Q17DD8 A0A182GK31 A0A1S4F713 A0A182F1D3 A0A336LD73 A0A182LMU2 V9IK84 A0A182KEL7 A0A182U044 A0A182V7Y3 A0A1S4HED3 A0A182RIR1 A0A084W5I9 A0A182Y0C0 E2BSL0 U5EWG4 A0A182XIW5 Q7Q0N5 A0A1S4H3F0 A0A0K8TPF4 A0A182I7B4 A0A182XNQ6 A0A182I0S9 K7J8Z0 A0A182N9R3 A0A182MN44 A0A182MZ66 A0A195EFZ2 A0A088A315 A0A182Q6K4 A0A084W7D9 A0A336KBV3 A0A182T2W5 A0A182QND5 A0A232EJ49 A0A182JQB1 A0A182FMX1 W5JA71 E2AC21 A0A182J7S8 A0A182V766 A0A084W7E0 A0A182KEE7 A0A2M4AGU2 A0A151I564 A0A2M4AH12 A0A0J7KHC8 A0A195CKM7 Q7PWM4 A0A2M3Z6V2 A0A182N8S8 Q16NY3 A0A182VSW2 A0A151WJM9 A0A182GCH0 A0A195F6Q3 A0A1S4H0D0 Q16RJ3 W5JRA5 A0A2M3Z2Q5 A0A2M3Z2U7 A0A182U7D5 A0A2M4BIX4 A0A2M4BIL8 A0A2M4BIM3
A0A194PMC0 A0A3S2M5M0 A0A0L7LCA7 A0A1L8E275 A0A1L8E2D3 A0A1B0D2V9 B0W7T9 A0A2M3Z3J8 A0A1Q3FF98 A0A1Q3FF50 A0A2M3Z3Q9 A0A182GK30 A0A2M4AH88 A0A2M4AH32 A0A023EW61 A0A182GCH1 A0A2M4BIR4 A0A2M4BII3 W5JRT3 B0W7U0 A0A2M4BIN3 A0A182SZG7 A0A182LXZ4 A0A1Q3F715 A0A1Q3F6X3 A0A182VUE3 A0A336N0K6 A0A182HCT7 A0A182KJU5 Q17DD8 A0A182GK31 A0A1S4F713 A0A182F1D3 A0A336LD73 A0A182LMU2 V9IK84 A0A182KEL7 A0A182U044 A0A182V7Y3 A0A1S4HED3 A0A182RIR1 A0A084W5I9 A0A182Y0C0 E2BSL0 U5EWG4 A0A182XIW5 Q7Q0N5 A0A1S4H3F0 A0A0K8TPF4 A0A182I7B4 A0A182XNQ6 A0A182I0S9 K7J8Z0 A0A182N9R3 A0A182MN44 A0A182MZ66 A0A195EFZ2 A0A088A315 A0A182Q6K4 A0A084W7D9 A0A336KBV3 A0A182T2W5 A0A182QND5 A0A232EJ49 A0A182JQB1 A0A182FMX1 W5JA71 E2AC21 A0A182J7S8 A0A182V766 A0A084W7E0 A0A182KEE7 A0A2M4AGU2 A0A151I564 A0A2M4AH12 A0A0J7KHC8 A0A195CKM7 Q7PWM4 A0A2M3Z6V2 A0A182N8S8 Q16NY3 A0A182VSW2 A0A151WJM9 A0A182GCH0 A0A195F6Q3 A0A1S4H0D0 Q16RJ3 W5JRA5 A0A2M3Z2Q5 A0A2M3Z2U7 A0A182U7D5 A0A2M4BIX4 A0A2M4BIL8 A0A2M4BIM3
PDB
4GDX
E-value=4.97432e-29,
Score=317
Ontologies
PATHWAY
GO
PANTHER
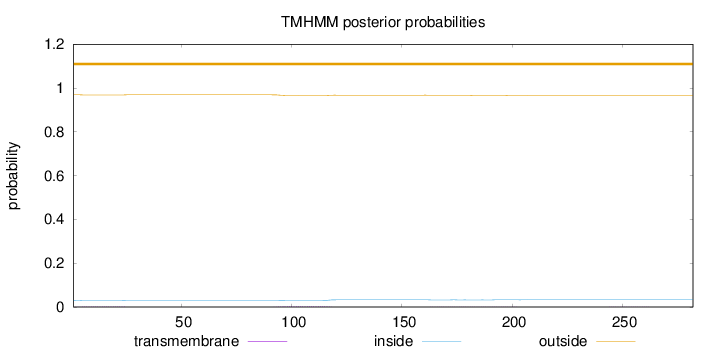
Topology
Length:
282
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15095
Exp number, first 60 AAs:
0.03634
Total prob of N-in:
0.03066
outside
1 - 282
Population Genetic Test Statistics
Pi
159.55214
Theta
2.976914
Tajima's D
-1.481258
CLR
193.677439
CSRT
0.0520473976301185
Interpretation
Uncertain
