Gene
KWMTBOMO04495
Pre Gene Modal
BGIBMGA005364
Annotation
PREDICTED:_uncharacterized_protein_LOC101744481_[Bombyx_mori]
Full name
Protein singed wings 2
Location in the cell
PlasmaMembrane Reliability : 2.333
Sequence
CDS
ATGCACGTGTTCTTGGGTCAGAATCCGTGGCGTTGTGACTGTCATTTCATACCCCGTTTTCAGGGACTCTTATTGAAGTACAAGAGGGTCATCAGGGATCTAACCGATATACGCTGCTCTAAGTCAAGCGATAAGAAAACATCGCTCGTACAAATCAGCACAATGCCACTCGGTAATGTTTGCAGCAATGAAACAGAGCTGGCAATTAGTCCGATTAACATTGTAAACATAGTTTTATTCGCATTAATAATGTTTATTCTAGCGGGCTAA
Protein
MHVFLGQNPWRCDCHFIPRFQGLLLKYKRVIRDLTDIRCSKSSDKKTSLVQISTMPLGNVCSNETELAISPINIVNIVLFALIMFILAG
Summary
Description
Has a role in the ecdysone induced cascade; probably indirect control of 'late' ecdysone genes.
Keywords
Complete proteome
Leucine-rich repeat
Reference proteome
Repeat
Signal
Feature
chain Protein singed wings 2
Uniprot
A0A1E1WKM4
A0A3S2LQ48
A0A2H1W280
A0A2A4JP72
A0A194QSA4
A0A194PN96
+ More
A0A0L7LMV5 A0A067RDZ9 A0A2J7RDB6 N6TJ47 K7ITT9 A0A195D470 A0A151X3M6 A0A195EFW6 A0A1Y1L253 A0A232FC61 E2CA86 A0A1B0D980 A0A195BQ55 A0A158NTL5 A0A1B6J9N9 A0A1B6FAT0 A0A2S2QCJ1 U4UD10 F4WQY7 A0A1Y1L275 A0A1Y1L0R1 A0A026VZE6 A0A195ES17 A0A1B6L5F7 J9LS61 E0VKW4 A0A1Y1KUX9 A0A088A1A5 A0A1L8DBV2 A0A0A9XE94 A0A0T6BDG7 D6WW75 A0A3Q0JEZ7 A0A0C9RHM2 A0A0C9QRS9 A0A310SBV1 A0A0K8WAZ6 A0A0M9AAK0 A0A1W4V0B6 A0A034W0K2 A0A1W4VDX3 A0A0A1WJR8 A0A034VXX9 A0A1A9VQB0 B3MIE4 B3NML7 Q28Z47 B4MPP0 A0A0L0BNM4 B4KMX3 A0A0J9U4T0 B4HMZ0 A0A1I8NX06 Q8SXT3 A0A1I8NX04 A0A1I8NX07 A0A1I8NX38 A0A0L7R5S5 A0A3B0IZ72 B4GIA5 A0A1A9YCZ0 A0A0R1DSY4 B4P555 B4LNC5 A0A0Q9W3W9 A0A1B6EGL5 A0A1B6DM74 A0A0P5GJC2 A0A0P4YWW5 A0A0P5LKC5 A0A0N8C925 A0A0P5ZJL1 A0A0P5WG67 A0A0M3QUS8 A0A0P6C508 A0A1B0C1U0 A0A2P8XWD8 W8BWS6 A0A182JFK5 B4J8L4 A0A084WDB6 A0A336M0B6 A0A0P6H9E6 B0WXK8 A0A182SBK3 A0A182G2P0 Q174X8 A0A182FHP4 W5JLW4 A0NEC5
A0A0L7LMV5 A0A067RDZ9 A0A2J7RDB6 N6TJ47 K7ITT9 A0A195D470 A0A151X3M6 A0A195EFW6 A0A1Y1L253 A0A232FC61 E2CA86 A0A1B0D980 A0A195BQ55 A0A158NTL5 A0A1B6J9N9 A0A1B6FAT0 A0A2S2QCJ1 U4UD10 F4WQY7 A0A1Y1L275 A0A1Y1L0R1 A0A026VZE6 A0A195ES17 A0A1B6L5F7 J9LS61 E0VKW4 A0A1Y1KUX9 A0A088A1A5 A0A1L8DBV2 A0A0A9XE94 A0A0T6BDG7 D6WW75 A0A3Q0JEZ7 A0A0C9RHM2 A0A0C9QRS9 A0A310SBV1 A0A0K8WAZ6 A0A0M9AAK0 A0A1W4V0B6 A0A034W0K2 A0A1W4VDX3 A0A0A1WJR8 A0A034VXX9 A0A1A9VQB0 B3MIE4 B3NML7 Q28Z47 B4MPP0 A0A0L0BNM4 B4KMX3 A0A0J9U4T0 B4HMZ0 A0A1I8NX06 Q8SXT3 A0A1I8NX04 A0A1I8NX07 A0A1I8NX38 A0A0L7R5S5 A0A3B0IZ72 B4GIA5 A0A1A9YCZ0 A0A0R1DSY4 B4P555 B4LNC5 A0A0Q9W3W9 A0A1B6EGL5 A0A1B6DM74 A0A0P5GJC2 A0A0P4YWW5 A0A0P5LKC5 A0A0N8C925 A0A0P5ZJL1 A0A0P5WG67 A0A0M3QUS8 A0A0P6C508 A0A1B0C1U0 A0A2P8XWD8 W8BWS6 A0A182JFK5 B4J8L4 A0A084WDB6 A0A336M0B6 A0A0P6H9E6 B0WXK8 A0A182SBK3 A0A182G2P0 Q174X8 A0A182FHP4 W5JLW4 A0NEC5
Pubmed
26354079
26227816
24845553
23537049
20075255
28004739
+ More
28648823 20798317 21347285 21719571 24508170 30249741 20566863 25401762 18362917 19820115 25348373 25830018 17994087 15632085 26108605 22936249 10731132 12537572 12537569 15189568 17550304 29403074 24495485 24438588 26483478 17510324 20920257 23761445 12364791 14747013 17210077
28648823 20798317 21347285 21719571 24508170 30249741 20566863 25401762 18362917 19820115 25348373 25830018 17994087 15632085 26108605 22936249 10731132 12537572 12537569 15189568 17550304 29403074 24495485 24438588 26483478 17510324 20920257 23761445 12364791 14747013 17210077
EMBL
GDQN01003638
JAT87416.1
RSAL01000032
RVE51526.1
ODYU01005754
SOQ46932.1
+ More
NWSH01000915 PCG73556.1 KQ461154 KPJ08403.1 KQ459599 KPI94463.1 JTDY01000535 KOB76760.1 KK852528 KDR21992.1 NEVH01005291 PNF38826.1 APGK01035612 KB740928 ENN77873.1 KQ976881 KYN07631.1 KQ982562 KYQ54860.1 KQ978983 KYN26762.1 GEZM01073287 GEZM01073281 JAV65187.1 NNAY01000490 OXU28028.1 GL453952 EFN75135.1 AJVK01004272 KQ976424 KYM88657.1 ADTU01026019 GECU01011814 JAS95892.1 GECZ01022477 JAS47292.1 GGMS01006262 MBY75465.1 KB632064 ERL88481.1 GL888275 EGI63500.1 GEZM01073284 GEZM01073280 JAV65207.1 GEZM01073283 GEZM01073282 JAV65196.1 KK107549 QOIP01000010 EZA49050.1 RLU17306.1 KQ981993 KYN30971.1 GEBQ01021092 JAT18885.1 ABLF02034555 DS235255 EEB14020.1 GEZM01073286 GEZM01073285 JAV65192.1 GFDF01010162 JAV03922.1 GBHO01025280 JAG18324.1 LJIG01001587 KRT85353.1 KQ971361 EFA09304.2 GBYB01006432 GBYB01006436 JAG76199.1 JAG76203.1 GBYB01006434 JAG76201.1 KQ764881 OAD54338.1 GDHF01004072 JAI48242.1 KQ435710 KOX79866.1 GAKP01011669 JAC47283.1 GBXI01015190 JAC99101.1 GAKP01011668 JAC47284.1 CH902619 EDV36992.2 CH954179 EDV54956.2 CM000071 EAL25768.3 CH963849 EDW74079.1 JRES01001589 KNC21690.1 CH933808 EDW09895.2 CM002911 KMY94655.1 CH480816 EDW48340.1 AE013599 AY084149 AAF57813.3 AAL89887.1 KQ414652 KOC66116.1 OUUW01000001 SPP73267.1 CH479183 EDW36225.1 CM000158 KRK00063.1 EDW91756.1 CH940648 EDW61077.2 KRF79763.1 GEDC01015292 GEDC01000212 JAS22006.1 JAS37086.1 GEDC01017727 GEDC01010524 JAS19571.1 JAS26774.1 GDIP01144302 GDIQ01246113 GDIQ01184678 LRGB01001005 JAJ79100.1 JAK05612.1 KZS14053.1 GDIP01225997 JAI97404.1 GDIQ01193269 JAK58456.1 GDIQ01102859 JAL48867.1 GDIP01042709 JAM61006.1 GDIP01099803 JAM03912.1 CP012524 ALC41189.1 GDIP01009083 JAM94632.1 JXJN01024172 PYGN01001247 PSN36318.1 GAMC01000795 JAC05761.1 CH916367 EDW01281.1 ATLV01022985 KE525339 KFB48210.1 UFQS01000257 UFQT01000257 SSX02072.1 SSX22449.1 GDIQ01034687 JAN60050.1 DS232168 EDS36538.1 JXUM01139872 KQ568969 KXJ68816.1 CH477404 EAT41653.1 ADMH02001054 ETN64293.1 AAAB01008960 EAU76444.2
NWSH01000915 PCG73556.1 KQ461154 KPJ08403.1 KQ459599 KPI94463.1 JTDY01000535 KOB76760.1 KK852528 KDR21992.1 NEVH01005291 PNF38826.1 APGK01035612 KB740928 ENN77873.1 KQ976881 KYN07631.1 KQ982562 KYQ54860.1 KQ978983 KYN26762.1 GEZM01073287 GEZM01073281 JAV65187.1 NNAY01000490 OXU28028.1 GL453952 EFN75135.1 AJVK01004272 KQ976424 KYM88657.1 ADTU01026019 GECU01011814 JAS95892.1 GECZ01022477 JAS47292.1 GGMS01006262 MBY75465.1 KB632064 ERL88481.1 GL888275 EGI63500.1 GEZM01073284 GEZM01073280 JAV65207.1 GEZM01073283 GEZM01073282 JAV65196.1 KK107549 QOIP01000010 EZA49050.1 RLU17306.1 KQ981993 KYN30971.1 GEBQ01021092 JAT18885.1 ABLF02034555 DS235255 EEB14020.1 GEZM01073286 GEZM01073285 JAV65192.1 GFDF01010162 JAV03922.1 GBHO01025280 JAG18324.1 LJIG01001587 KRT85353.1 KQ971361 EFA09304.2 GBYB01006432 GBYB01006436 JAG76199.1 JAG76203.1 GBYB01006434 JAG76201.1 KQ764881 OAD54338.1 GDHF01004072 JAI48242.1 KQ435710 KOX79866.1 GAKP01011669 JAC47283.1 GBXI01015190 JAC99101.1 GAKP01011668 JAC47284.1 CH902619 EDV36992.2 CH954179 EDV54956.2 CM000071 EAL25768.3 CH963849 EDW74079.1 JRES01001589 KNC21690.1 CH933808 EDW09895.2 CM002911 KMY94655.1 CH480816 EDW48340.1 AE013599 AY084149 AAF57813.3 AAL89887.1 KQ414652 KOC66116.1 OUUW01000001 SPP73267.1 CH479183 EDW36225.1 CM000158 KRK00063.1 EDW91756.1 CH940648 EDW61077.2 KRF79763.1 GEDC01015292 GEDC01000212 JAS22006.1 JAS37086.1 GEDC01017727 GEDC01010524 JAS19571.1 JAS26774.1 GDIP01144302 GDIQ01246113 GDIQ01184678 LRGB01001005 JAJ79100.1 JAK05612.1 KZS14053.1 GDIP01225997 JAI97404.1 GDIQ01193269 JAK58456.1 GDIQ01102859 JAL48867.1 GDIP01042709 JAM61006.1 GDIP01099803 JAM03912.1 CP012524 ALC41189.1 GDIP01009083 JAM94632.1 JXJN01024172 PYGN01001247 PSN36318.1 GAMC01000795 JAC05761.1 CH916367 EDW01281.1 ATLV01022985 KE525339 KFB48210.1 UFQS01000257 UFQT01000257 SSX02072.1 SSX22449.1 GDIQ01034687 JAN60050.1 DS232168 EDS36538.1 JXUM01139872 KQ568969 KXJ68816.1 CH477404 EAT41653.1 ADMH02001054 ETN64293.1 AAAB01008960 EAU76444.2
Proteomes
UP000283053
UP000218220
UP000053240
UP000053268
UP000037510
UP000027135
+ More
UP000235965 UP000019118 UP000002358 UP000078542 UP000075809 UP000078492 UP000215335 UP000008237 UP000092462 UP000078540 UP000005205 UP000030742 UP000007755 UP000053097 UP000279307 UP000078541 UP000007819 UP000009046 UP000005203 UP000007266 UP000079169 UP000053105 UP000192221 UP000078200 UP000007801 UP000008711 UP000001819 UP000007798 UP000037069 UP000009192 UP000001292 UP000095300 UP000000803 UP000053825 UP000268350 UP000008744 UP000092443 UP000002282 UP000008792 UP000076858 UP000092553 UP000092460 UP000245037 UP000075880 UP000001070 UP000030765 UP000002320 UP000075901 UP000069940 UP000249989 UP000008820 UP000069272 UP000000673 UP000007062
UP000235965 UP000019118 UP000002358 UP000078542 UP000075809 UP000078492 UP000215335 UP000008237 UP000092462 UP000078540 UP000005205 UP000030742 UP000007755 UP000053097 UP000279307 UP000078541 UP000007819 UP000009046 UP000005203 UP000007266 UP000079169 UP000053105 UP000192221 UP000078200 UP000007801 UP000008711 UP000001819 UP000007798 UP000037069 UP000009192 UP000001292 UP000095300 UP000000803 UP000053825 UP000268350 UP000008744 UP000092443 UP000002282 UP000008792 UP000076858 UP000092553 UP000092460 UP000245037 UP000075880 UP000001070 UP000030765 UP000002320 UP000075901 UP000069940 UP000249989 UP000008820 UP000069272 UP000000673 UP000007062
PRIDE
Pfam
PF13855 LRR_8
Interpro
Gene 3D
ProteinModelPortal
A0A1E1WKM4
A0A3S2LQ48
A0A2H1W280
A0A2A4JP72
A0A194QSA4
A0A194PN96
+ More
A0A0L7LMV5 A0A067RDZ9 A0A2J7RDB6 N6TJ47 K7ITT9 A0A195D470 A0A151X3M6 A0A195EFW6 A0A1Y1L253 A0A232FC61 E2CA86 A0A1B0D980 A0A195BQ55 A0A158NTL5 A0A1B6J9N9 A0A1B6FAT0 A0A2S2QCJ1 U4UD10 F4WQY7 A0A1Y1L275 A0A1Y1L0R1 A0A026VZE6 A0A195ES17 A0A1B6L5F7 J9LS61 E0VKW4 A0A1Y1KUX9 A0A088A1A5 A0A1L8DBV2 A0A0A9XE94 A0A0T6BDG7 D6WW75 A0A3Q0JEZ7 A0A0C9RHM2 A0A0C9QRS9 A0A310SBV1 A0A0K8WAZ6 A0A0M9AAK0 A0A1W4V0B6 A0A034W0K2 A0A1W4VDX3 A0A0A1WJR8 A0A034VXX9 A0A1A9VQB0 B3MIE4 B3NML7 Q28Z47 B4MPP0 A0A0L0BNM4 B4KMX3 A0A0J9U4T0 B4HMZ0 A0A1I8NX06 Q8SXT3 A0A1I8NX04 A0A1I8NX07 A0A1I8NX38 A0A0L7R5S5 A0A3B0IZ72 B4GIA5 A0A1A9YCZ0 A0A0R1DSY4 B4P555 B4LNC5 A0A0Q9W3W9 A0A1B6EGL5 A0A1B6DM74 A0A0P5GJC2 A0A0P4YWW5 A0A0P5LKC5 A0A0N8C925 A0A0P5ZJL1 A0A0P5WG67 A0A0M3QUS8 A0A0P6C508 A0A1B0C1U0 A0A2P8XWD8 W8BWS6 A0A182JFK5 B4J8L4 A0A084WDB6 A0A336M0B6 A0A0P6H9E6 B0WXK8 A0A182SBK3 A0A182G2P0 Q174X8 A0A182FHP4 W5JLW4 A0NEC5
A0A0L7LMV5 A0A067RDZ9 A0A2J7RDB6 N6TJ47 K7ITT9 A0A195D470 A0A151X3M6 A0A195EFW6 A0A1Y1L253 A0A232FC61 E2CA86 A0A1B0D980 A0A195BQ55 A0A158NTL5 A0A1B6J9N9 A0A1B6FAT0 A0A2S2QCJ1 U4UD10 F4WQY7 A0A1Y1L275 A0A1Y1L0R1 A0A026VZE6 A0A195ES17 A0A1B6L5F7 J9LS61 E0VKW4 A0A1Y1KUX9 A0A088A1A5 A0A1L8DBV2 A0A0A9XE94 A0A0T6BDG7 D6WW75 A0A3Q0JEZ7 A0A0C9RHM2 A0A0C9QRS9 A0A310SBV1 A0A0K8WAZ6 A0A0M9AAK0 A0A1W4V0B6 A0A034W0K2 A0A1W4VDX3 A0A0A1WJR8 A0A034VXX9 A0A1A9VQB0 B3MIE4 B3NML7 Q28Z47 B4MPP0 A0A0L0BNM4 B4KMX3 A0A0J9U4T0 B4HMZ0 A0A1I8NX06 Q8SXT3 A0A1I8NX04 A0A1I8NX07 A0A1I8NX38 A0A0L7R5S5 A0A3B0IZ72 B4GIA5 A0A1A9YCZ0 A0A0R1DSY4 B4P555 B4LNC5 A0A0Q9W3W9 A0A1B6EGL5 A0A1B6DM74 A0A0P5GJC2 A0A0P4YWW5 A0A0P5LKC5 A0A0N8C925 A0A0P5ZJL1 A0A0P5WG67 A0A0M3QUS8 A0A0P6C508 A0A1B0C1U0 A0A2P8XWD8 W8BWS6 A0A182JFK5 B4J8L4 A0A084WDB6 A0A336M0B6 A0A0P6H9E6 B0WXK8 A0A182SBK3 A0A182G2P0 Q174X8 A0A182FHP4 W5JLW4 A0NEC5
PDB
3TWI
E-value=0.0820525,
Score=77
Ontologies
PANTHER
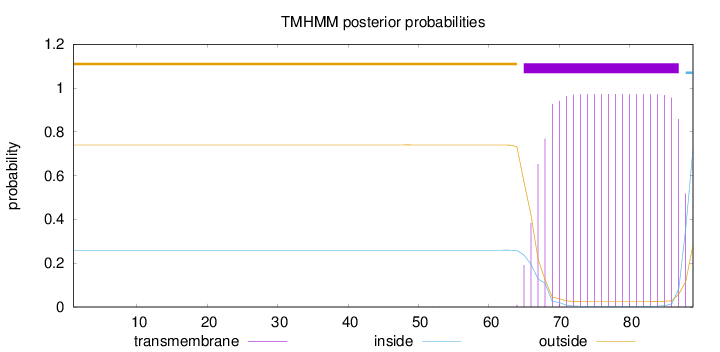
Topology
Length:
89
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.78442
Exp number, first 60 AAs:
0.00323
Total prob of N-in:
0.25961
outside
1 - 64
TMhelix
65 - 87
inside
88 - 89
Population Genetic Test Statistics
Pi
202.844071
Theta
180.050386
Tajima's D
0.815446
CLR
1.782642
CSRT
0.606519674016299
Interpretation
Uncertain
