Gene
KWMTBOMO04491
Pre Gene Modal
BGIBMGA005365
Annotation
PREDICTED:_uncharacterized_protein_LOC101744340_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.914 Nuclear Reliability : 1.413
Sequence
CDS
ATGTACGTCTTAAAAGACTTTCAAGACGTTTCGAACAACAGAACTTTTCTGTTCCTAATAGTGTTGCTATCGTTAAAGCTGTGTTGTCCCCAAGACTTGAGCGAAGATTTCGTGAACGAGGATAGTGCAGACGTCGTGTCAGACTTAAGTATGGTAGTTGAAACGGTAAAACTGTCAAGGAGCGTTGTGGGATCGCCCTGTAGAATGAGCGAGTTGCTATGCGACACTGGGCAATGTATATCTTTGGATAAGTACTGCAACGGAGAAGACGATTGTGGAGACAAGAGCGACGAACCCAAGAGCTGTACACCGTGTAACAGAACATACATGGGTGATGTGGGTCGAACTTACGAGTTGGAAGTACGACGGCCTCGGGAAGATCATCTGCCCTTCGTCTGCCACCTCAACTTCACAGCAACAGGAGGGAACTACGGAGATATAATTCAGTTGACATTTGACACGTTTACCGTCGGCAAATTCGTGTCGTTCACGTCAGACGGTTGTCCGGATGGGCACATGACAATTGTGGAGAGAAGTCCGTCACCGCCCACCGGTCAGTGGTGCGGCTCTGCTTGGGGTTACACAGTGTACTTCAGCGAATCCGATTCCATAAACATGACTCTCCGACTGGACAGGCTAAGTCAACAGGGTGTTGGCTACAATTTCGACTTCAAGTTGGCTTACAAGTTTTTGAGGCGAAGCGAAGCTCGGCTGCGATACGGAAATGCGACTGTAGGGGCGTGGAGGGGAGAGAGGATCCCTGGTACATACTGTGACCGCATCCTTAGCGACTGTGATCTGAGGGCCTGCCGCATACAGTCACCAAACTTTCCTGGAGTGTATCCTCGCAACGCTACTTGTACATACCGAATCGAACATACTAAGATACCAGCCGATAAACATGTGCTGCTGGCAGTTCGACAGACGAACAGCCACAAAATACATATCAAAGACCAAATAGTGAAGTACGATAGAAGTCAACGTGTTTTAAAAATATGGGATCAATGTAACGTAGTTCAGGATTACTTAACAGTTTGGGATGGATCCACTAGAGATTCGCCAGTATTAGTGCGGCTTTGTGGAGGAGATGCTGTACCGGATATTATCAGTAGAGGTCCGAACATGTTACTGGAGTTTCATACGTCACCTTACGATAATCCATTTCATCCGGTACCGTTGAGCTATTTACCTGGGTTTGAATTGGAAGTTCAGGTTTTGTATGTGGACAAAGATTCTCATTCGTACGTGCGCACAGACGGCAGGTGCCGTTTTGTTTTACGTTCTTCTGACAAAACTAGCGGAATACTCAGGAATCCAAGACACTCGCTACCTCCAAATACATCGTGCGTCTACTATTTTCAGGGTCGTTCAAATGAAATAGTATGGGTGTCGTTTGTGAAATATCATTCGGCTGGAACTGAGCCGGCAGGTTTTGATCAGCAGCAAGATTGTTCGTCTCAATTGACTATTTGGGACGGGGCTGCCCCCGACGCCGACCTCGATAGGAAGCTGGATATGACGGATCAGAGATCTCTACTTGGTTCGTTCTGTCGTGAGGAATCTCCACGACTATGCGATCATGCGCTTTTGTCAAATTCTACAAGGGCGACGCGGCCTTGCGCTCCTTCGGAGAGCTACATCACTACCGGACCTGCTTTGACAATTCTTCAGGAACTACGTCAAGGGTCCGCATTATATCCGGTATCGTTTTTACTACGGTATGAATTCGTCGATGTGAGTGAACAAGGACAGCCTTTGACAGATTCTAAGTCCGCTTGCGATAGAGTCTTTAAATCAGCTCTAACATATTCTGGAAGGTTCCAGGCCCCACGAGCCATATTTTACTATGGACGCGGCGGAGCACAAAACTTAACCTGTATTTTAAGATTTGAAGCAAAACAGGGCGAGAGGGTTCAGTTGACCTTTTTGAATACCTTTTTTGGGAACAAACATTGCGGTACACACAAAGATTCCCGCACAAGTCGCTGGAAATGCGATAGGCCTCCGAGAAGAACTATCGGGGGTGAAGGGTTTGCTCAGATCATTATCACGGAGTTCCCTTGGGTGGGAGTTCCGATTCAACGCGACTGTATTTGCACAAATCGATCAGAGCCTATAAATATACACACATTAACTGCACCAGTAGTTGAAGTAAATTTTACTGTAACAATGATGAATATTACAGAGGATTATGACGATTTTGCCTTTGAGGGAGAATATAAATTTATTCCGACCGGTCCTGGTGATGATACTGTATGTTCTACTGGTTGGGGTGAAAGACGCGTTAGAGGAAGCAGTGGAGAAATTCGGCTCTATGATAAAAGACAAATGACTATAGTGCCAGAGATTGTCGGTGACAGGAACGTTATATCAGAGAGCGTAAGAGCAGAAGTTGCTTGTGTTCATAGACCATGGTTAATTGAACCTGGAGGTGATGATATTTCCCCGGTTCAGGGTAGATATTTATATGTAAAGGTTCCAGGGTATGAGATAACGTCTTCATCGCCGTTCTGCTCTACTCCAAACAGACTTTTTATTTACGAAGCTCGTGATACAACTCGATCCAAAGAGATATGTCCTGATATAAACAACAACTTGGAATTATTTTCTCCTGACTGGAAATCTTCTGAGACTCCATTAGAAACATCTCTAAAACCTCACGCACGAAGCTACGTTATTGACTTTCTTCAGCACGAACAAGCAGATTATTCAATAAAGTGGATTGAGCTGATGAAGAAACCTAACTTTGATCATGACCCTGACACAAATGTCTTACCTGAAGCCGTTTCATTTGATTGTCTCCACAGCTGTCCCGAATTAAATGCGTGTATTCCTATTGCACTGTGGTGTGATGGAAGCGCCCACTGTCCTTCAGGTTACGACGAAGACGATTCCAACTGCTCGTTCAGATTATCATTGCCTCCTCCGTACATTGTGGCCGCTGGAGGCGTGGCTTTGCTGATATGTGCAGTTCTAATAGCTTTCTGTGCTTGTAAAAAGCGAAAACGAAAGGACAAAGAATTTAAAGCTAGGCTAGATGGAGCAATACCTACTGAAGAAAGGCCCTTTGACAGAGCTAAAGCTAATGGAGTCCCCGAAGCGTCACGACAGTATGCCACAGTCCAGAAATATGCGACAATAGACAAATATTCTTTGAGCCAGAAATATAGTGCTGGCTTAAACGATGCGAGGTATTACGACGAAGTAGCACAAAGAGATAAGCTGGCAGATACAAGGTACGCTAGTCTGGGAAGGGGAAGCCGAAATATGCGCTACGAAAATTCTAGAGGTAACGGTTCAAGAAGAGCAATGCCTGATTTAGGTTATCCCGACTTGAAAGATGGTTTTTGTTGA
Protein
MYVLKDFQDVSNNRTFLFLIVLLSLKLCCPQDLSEDFVNEDSADVVSDLSMVVETVKLSRSVVGSPCRMSELLCDTGQCISLDKYCNGEDDCGDKSDEPKSCTPCNRTYMGDVGRTYELEVRRPREDHLPFVCHLNFTATGGNYGDIIQLTFDTFTVGKFVSFTSDGCPDGHMTIVERSPSPPTGQWCGSAWGYTVYFSESDSINMTLRLDRLSQQGVGYNFDFKLAYKFLRRSEARLRYGNATVGAWRGERIPGTYCDRILSDCDLRACRIQSPNFPGVYPRNATCTYRIEHTKIPADKHVLLAVRQTNSHKIHIKDQIVKYDRSQRVLKIWDQCNVVQDYLTVWDGSTRDSPVLVRLCGGDAVPDIISRGPNMLLEFHTSPYDNPFHPVPLSYLPGFELEVQVLYVDKDSHSYVRTDGRCRFVLRSSDKTSGILRNPRHSLPPNTSCVYYFQGRSNEIVWVSFVKYHSAGTEPAGFDQQQDCSSQLTIWDGAAPDADLDRKLDMTDQRSLLGSFCREESPRLCDHALLSNSTRATRPCAPSESYITTGPALTILQELRQGSALYPVSFLLRYEFVDVSEQGQPLTDSKSACDRVFKSALTYSGRFQAPRAIFYYGRGGAQNLTCILRFEAKQGERVQLTFLNTFFGNKHCGTHKDSRTSRWKCDRPPRRTIGGEGFAQIIITEFPWVGVPIQRDCICTNRSEPINIHTLTAPVVEVNFTVTMMNITEDYDDFAFEGEYKFIPTGPGDDTVCSTGWGERRVRGSSGEIRLYDKRQMTIVPEIVGDRNVISESVRAEVACVHRPWLIEPGGDDISPVQGRYLYVKVPGYEITSSSPFCSTPNRLFIYEARDTTRSKEICPDINNNLELFSPDWKSSETPLETSLKPHARSYVIDFLQHEQADYSIKWIELMKKPNFDHDPDTNVLPEAVSFDCLHSCPELNACIPIALWCDGSAHCPSGYDEDDSNCSFRLSLPPPYIVAAGGVALLICAVLIAFCACKKRKRKDKEFKARLDGAIPTEERPFDRAKANGVPEASRQYATVQKYATIDKYSLSQKYSAGLNDARYYDEVAQRDKLADTRYASLGRGSRNMRYENSRGNGSRRAMPDLGYPDLKDGFC
Summary
Uniprot
H9J773
A0A3S2LUX9
A0A212EIH7
A0A194PLZ7
A0A2A4J7I9
A0A2H1WIR5
+ More
A0A194QTS9 A0A0L7L3D5 A0A026VZ22 A0A195D421 A0A195EFU4 E2AVC0 A0A151X384 A0A0L7QU87 A0A195BQ69 F4WQY5 A0A154P0K5 A0A0M9A387 J9JL19 A0A2S2R1E3 A0A195ESW0 A0A0J7L4Y6 D6WXF4 A0A2A3EN67 E0VKW7 A0A088A1X7 A0A182JIR3 A0A182P1U2 A0A182QA36 A0A182KVE5 A0A336MTI9 A0A088A128 A0A158NDI5 A0A2J7RGS4 A0A067R9X5 A0A139WBM9 A0A3L8DN15 E0V9K2 B1B558 A0A194QWE6 A0A182YIZ7 A0A195CEV3 F4W516 A0A3S2M9Y6 A0A195D9W5 A0A182UTJ4 Q17NX4 E0W0Y7 A0A2P8ZKH1 A0A195BQ33 A0A1W4WIT4 A0A1B0CN39 A0A0L7QUS5 A0A154PRJ0 T1H809 A0A212F0K8 T1JCM1 D6WXQ1 K7J771 A0A088A0Z9 Q7Q7T1 A0A151IBQ7 A0A195B511 F4WHN2 A0A2S2QIH8 A0A2W1BRG3 J9LBR5 A0A2A4K9S5 A0A212FJW9 A0A151WIT4 A0A2H1VYU1 A0A0C9RAS7 N6T4A6 A0A2A3ELJ0 T1JBM9 A0A0N0U4U8 W8C8T0 E9IIH7 W8BX11 A0A151JW37 A0A0L7QUP5 E2BJM2 A0A158NM50 E2B1T4 A0A0C9R1A0 J9KQ07 A0A2S2PMN9 A0A194PWI1 A0A067R7E6
A0A194QTS9 A0A0L7L3D5 A0A026VZ22 A0A195D421 A0A195EFU4 E2AVC0 A0A151X384 A0A0L7QU87 A0A195BQ69 F4WQY5 A0A154P0K5 A0A0M9A387 J9JL19 A0A2S2R1E3 A0A195ESW0 A0A0J7L4Y6 D6WXF4 A0A2A3EN67 E0VKW7 A0A088A1X7 A0A182JIR3 A0A182P1U2 A0A182QA36 A0A182KVE5 A0A336MTI9 A0A088A128 A0A158NDI5 A0A2J7RGS4 A0A067R9X5 A0A139WBM9 A0A3L8DN15 E0V9K2 B1B558 A0A194QWE6 A0A182YIZ7 A0A195CEV3 F4W516 A0A3S2M9Y6 A0A195D9W5 A0A182UTJ4 Q17NX4 E0W0Y7 A0A2P8ZKH1 A0A195BQ33 A0A1W4WIT4 A0A1B0CN39 A0A0L7QUS5 A0A154PRJ0 T1H809 A0A212F0K8 T1JCM1 D6WXQ1 K7J771 A0A088A0Z9 Q7Q7T1 A0A151IBQ7 A0A195B511 F4WHN2 A0A2S2QIH8 A0A2W1BRG3 J9LBR5 A0A2A4K9S5 A0A212FJW9 A0A151WIT4 A0A2H1VYU1 A0A0C9RAS7 N6T4A6 A0A2A3ELJ0 T1JBM9 A0A0N0U4U8 W8C8T0 E9IIH7 W8BX11 A0A151JW37 A0A0L7QUP5 E2BJM2 A0A158NM50 E2B1T4 A0A0C9R1A0 J9KQ07 A0A2S2PMN9 A0A194PWI1 A0A067R7E6
Pubmed
EMBL
BABH01013734
RSAL01000196
RVE44555.1
AGBW02014633
OWR41289.1
KQ459599
+ More
KPI94461.1 NWSH01002522 PCG68087.1 ODYU01008937 SOQ52959.1 KQ461154 KPJ08405.1 JTDY01003175 KOB70013.1 KK107549 QOIP01000010 EZA49053.1 RLU17780.1 KQ976881 KYN07633.1 KQ978983 KYN26764.1 GL443064 EFN62616.1 KQ982562 KYQ54863.1 KQ414735 KOC62222.1 KQ976424 KYM88655.1 GL888275 EGI63498.1 KQ434792 KZC05459.1 KQ435776 KOX74929.1 ABLF02029318 ABLF02029319 ABLF02029320 ABLF02029321 ABLF02029322 ABLF02033880 ABLF02033883 ABLF02033885 ABLF02033887 ABLF02045612 GGMS01014616 MBY83819.1 KQ981993 KYN30969.1 LBMM01000589 KMQ98012.1 KQ971361 EFA07989.2 KZ288215 PBC32646.1 DS235255 EEB14023.1 AXCN02001812 UFQT01001887 SSX32093.1 ADTU01012606 ADTU01012607 ADTU01012608 ADTU01012609 ADTU01012610 ADTU01012611 NEVH01003752 PNF40041.1 KK852649 KDR19489.1 KQ971372 KYB25368.1 QOIP01000006 RLU21218.1 DS234994 EEB10058.1 AB360591 BAG12564.1 KQ461175 KPJ07891.1 KQ977827 KYM99419.1 GL887596 EGI70712.1 RSAL01000003 RVE54804.1 KQ981082 KYN09652.1 CH477195 EAT48424.1 DS235863 EEB19292.1 PYGN01000029 PSN56998.1 KYM88248.1 AJWK01019761 AJWK01019762 AJWK01019763 KOC62226.1 KQ435078 KZC14511.1 ACPB03006880 ACPB03006881 ACPB03006882 ACPB03006883 AGBW02011082 OWR47234.1 JH432065 KQ971362 EFA08886.1 AAAB01008951 EAA10533.4 KQ978079 KYM97151.1 KQ976604 KYM79365.1 GL888167 EGI66239.1 GGMS01008308 MBY77511.1 KZ149998 PZC75360.1 ABLF02039535 ABLF02039539 ABLF02050433 NWSH01000014 PCG80829.1 AGBW02008215 OWR54000.1 KQ983075 KYQ47758.1 ODYU01005285 SOQ45988.1 GBYB01010192 JAG79959.1 APGK01044261 APGK01044262 APGK01044263 KB741022 ENN74974.1 PBC32588.1 JH432011 KQ435821 KOX72482.1 GAMC01002774 JAC03782.1 GL763491 EFZ19623.1 GAMC01002773 JAC03783.1 KQ981663 KYN38502.1 KOC62299.1 GL448604 EFN84080.1 ADTU01020256 ADTU01020257 GL444956 EFN60355.1 GBYB01001735 JAG71502.1 ABLF02027959 GGMR01018035 MBY30654.1 KQ459590 KPI97119.1 KK852687 KDR18394.1
KPI94461.1 NWSH01002522 PCG68087.1 ODYU01008937 SOQ52959.1 KQ461154 KPJ08405.1 JTDY01003175 KOB70013.1 KK107549 QOIP01000010 EZA49053.1 RLU17780.1 KQ976881 KYN07633.1 KQ978983 KYN26764.1 GL443064 EFN62616.1 KQ982562 KYQ54863.1 KQ414735 KOC62222.1 KQ976424 KYM88655.1 GL888275 EGI63498.1 KQ434792 KZC05459.1 KQ435776 KOX74929.1 ABLF02029318 ABLF02029319 ABLF02029320 ABLF02029321 ABLF02029322 ABLF02033880 ABLF02033883 ABLF02033885 ABLF02033887 ABLF02045612 GGMS01014616 MBY83819.1 KQ981993 KYN30969.1 LBMM01000589 KMQ98012.1 KQ971361 EFA07989.2 KZ288215 PBC32646.1 DS235255 EEB14023.1 AXCN02001812 UFQT01001887 SSX32093.1 ADTU01012606 ADTU01012607 ADTU01012608 ADTU01012609 ADTU01012610 ADTU01012611 NEVH01003752 PNF40041.1 KK852649 KDR19489.1 KQ971372 KYB25368.1 QOIP01000006 RLU21218.1 DS234994 EEB10058.1 AB360591 BAG12564.1 KQ461175 KPJ07891.1 KQ977827 KYM99419.1 GL887596 EGI70712.1 RSAL01000003 RVE54804.1 KQ981082 KYN09652.1 CH477195 EAT48424.1 DS235863 EEB19292.1 PYGN01000029 PSN56998.1 KYM88248.1 AJWK01019761 AJWK01019762 AJWK01019763 KOC62226.1 KQ435078 KZC14511.1 ACPB03006880 ACPB03006881 ACPB03006882 ACPB03006883 AGBW02011082 OWR47234.1 JH432065 KQ971362 EFA08886.1 AAAB01008951 EAA10533.4 KQ978079 KYM97151.1 KQ976604 KYM79365.1 GL888167 EGI66239.1 GGMS01008308 MBY77511.1 KZ149998 PZC75360.1 ABLF02039535 ABLF02039539 ABLF02050433 NWSH01000014 PCG80829.1 AGBW02008215 OWR54000.1 KQ983075 KYQ47758.1 ODYU01005285 SOQ45988.1 GBYB01010192 JAG79959.1 APGK01044261 APGK01044262 APGK01044263 KB741022 ENN74974.1 PBC32588.1 JH432011 KQ435821 KOX72482.1 GAMC01002774 JAC03782.1 GL763491 EFZ19623.1 GAMC01002773 JAC03783.1 KQ981663 KYN38502.1 KOC62299.1 GL448604 EFN84080.1 ADTU01020256 ADTU01020257 GL444956 EFN60355.1 GBYB01001735 JAG71502.1 ABLF02027959 GGMR01018035 MBY30654.1 KQ459590 KPI97119.1 KK852687 KDR18394.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000218220
UP000053240
+ More
UP000037510 UP000053097 UP000279307 UP000078542 UP000078492 UP000000311 UP000075809 UP000053825 UP000078540 UP000007755 UP000076502 UP000053105 UP000007819 UP000078541 UP000036403 UP000007266 UP000242457 UP000009046 UP000005203 UP000075880 UP000075885 UP000075886 UP000075882 UP000005205 UP000235965 UP000027135 UP000076408 UP000075903 UP000008820 UP000245037 UP000192223 UP000092461 UP000015103 UP000002358 UP000007062 UP000019118 UP000008237
UP000037510 UP000053097 UP000279307 UP000078542 UP000078492 UP000000311 UP000075809 UP000053825 UP000078540 UP000007755 UP000076502 UP000053105 UP000007819 UP000078541 UP000036403 UP000007266 UP000242457 UP000009046 UP000005203 UP000075880 UP000075885 UP000075886 UP000075882 UP000005205 UP000235965 UP000027135 UP000076408 UP000075903 UP000008820 UP000245037 UP000192223 UP000092461 UP000015103 UP000002358 UP000007062 UP000019118 UP000008237
Interpro
Gene 3D
ProteinModelPortal
H9J773
A0A3S2LUX9
A0A212EIH7
A0A194PLZ7
A0A2A4J7I9
A0A2H1WIR5
+ More
A0A194QTS9 A0A0L7L3D5 A0A026VZ22 A0A195D421 A0A195EFU4 E2AVC0 A0A151X384 A0A0L7QU87 A0A195BQ69 F4WQY5 A0A154P0K5 A0A0M9A387 J9JL19 A0A2S2R1E3 A0A195ESW0 A0A0J7L4Y6 D6WXF4 A0A2A3EN67 E0VKW7 A0A088A1X7 A0A182JIR3 A0A182P1U2 A0A182QA36 A0A182KVE5 A0A336MTI9 A0A088A128 A0A158NDI5 A0A2J7RGS4 A0A067R9X5 A0A139WBM9 A0A3L8DN15 E0V9K2 B1B558 A0A194QWE6 A0A182YIZ7 A0A195CEV3 F4W516 A0A3S2M9Y6 A0A195D9W5 A0A182UTJ4 Q17NX4 E0W0Y7 A0A2P8ZKH1 A0A195BQ33 A0A1W4WIT4 A0A1B0CN39 A0A0L7QUS5 A0A154PRJ0 T1H809 A0A212F0K8 T1JCM1 D6WXQ1 K7J771 A0A088A0Z9 Q7Q7T1 A0A151IBQ7 A0A195B511 F4WHN2 A0A2S2QIH8 A0A2W1BRG3 J9LBR5 A0A2A4K9S5 A0A212FJW9 A0A151WIT4 A0A2H1VYU1 A0A0C9RAS7 N6T4A6 A0A2A3ELJ0 T1JBM9 A0A0N0U4U8 W8C8T0 E9IIH7 W8BX11 A0A151JW37 A0A0L7QUP5 E2BJM2 A0A158NM50 E2B1T4 A0A0C9R1A0 J9KQ07 A0A2S2PMN9 A0A194PWI1 A0A067R7E6
A0A194QTS9 A0A0L7L3D5 A0A026VZ22 A0A195D421 A0A195EFU4 E2AVC0 A0A151X384 A0A0L7QU87 A0A195BQ69 F4WQY5 A0A154P0K5 A0A0M9A387 J9JL19 A0A2S2R1E3 A0A195ESW0 A0A0J7L4Y6 D6WXF4 A0A2A3EN67 E0VKW7 A0A088A1X7 A0A182JIR3 A0A182P1U2 A0A182QA36 A0A182KVE5 A0A336MTI9 A0A088A128 A0A158NDI5 A0A2J7RGS4 A0A067R9X5 A0A139WBM9 A0A3L8DN15 E0V9K2 B1B558 A0A194QWE6 A0A182YIZ7 A0A195CEV3 F4W516 A0A3S2M9Y6 A0A195D9W5 A0A182UTJ4 Q17NX4 E0W0Y7 A0A2P8ZKH1 A0A195BQ33 A0A1W4WIT4 A0A1B0CN39 A0A0L7QUS5 A0A154PRJ0 T1H809 A0A212F0K8 T1JCM1 D6WXQ1 K7J771 A0A088A0Z9 Q7Q7T1 A0A151IBQ7 A0A195B511 F4WHN2 A0A2S2QIH8 A0A2W1BRG3 J9LBR5 A0A2A4K9S5 A0A212FJW9 A0A151WIT4 A0A2H1VYU1 A0A0C9RAS7 N6T4A6 A0A2A3ELJ0 T1JBM9 A0A0N0U4U8 W8C8T0 E9IIH7 W8BX11 A0A151JW37 A0A0L7QUP5 E2BJM2 A0A158NM50 E2B1T4 A0A0C9R1A0 J9KQ07 A0A2S2PMN9 A0A194PWI1 A0A067R7E6
PDB
6H04
E-value=0.00317384,
Score=100
Ontologies
GO
Topology
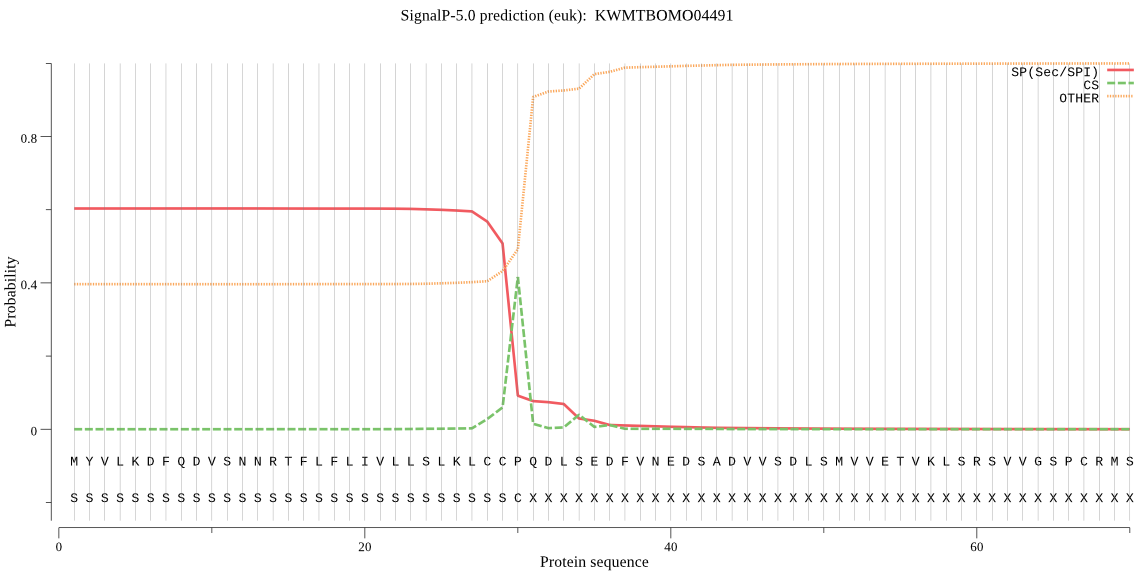
SignalP
Position: 1 - 30,
Likelihood: 0.604262
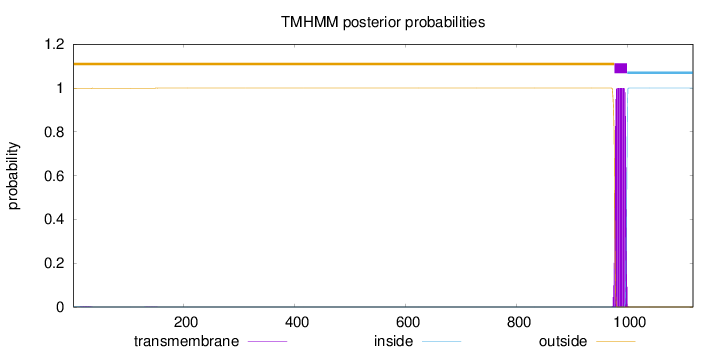
Length:
1117
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.71448
Exp number, first 60 AAs:
0.03988
Total prob of N-in:
0.00238
outside
1 - 975
TMhelix
976 - 998
inside
999 - 1117
Population Genetic Test Statistics
Pi
215.793349
Theta
187.804019
Tajima's D
0.78088
CLR
0.232045
CSRT
0.591770411479426
Interpretation
Uncertain
