Gene
KWMTBOMO04489 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005444
Annotation
Uncharacterized_protein_OBRU01_00156_[Operophtera_brumata]
Location in the cell
Mitochondrial Reliability : 2.314
Sequence
CDS
ATGGTGTCGTTAACTTCTAAATATATACTTTCAACGATATGTAAATCAAATAGCATTAGGGCATTATCTACAACGTATTCAAGAAATGCGAAAGTGACTTTCGATTGGGTGGACCCATTCAATTTAGATGGACAACTCCATGACGACGAAAAAGCAGTGCGAGATTCATTCAGGGCCTACTGTAATGAAAAGTTATTACCTCGTGTGGTTGAGGCAAATAGGAATGAGGTGTTCCATAGAGAAATCTATAATGAACTCGGTGAATTAGGCGCTCTGGGTTGTACCATTAAAGGTTACGGATGCGCTGGAGTTTCTTACGTTACGTATGGTCTCATTACAAGAGAACTAGATGGCGTTGACTCCTCTTATAGATCAGCGATGAGTGTGCAGAGTAGTTTGGCAATGGGTTCGATTTATATGTACGGAACTGAAGACCAGAAACAGAAATACTTACCACGCATGGCTACAGGTGAATTAATTGGTTGTTTTGGGTTAACGGAACCCAATTTCGGAAGTGATGCTGGAGGCTTAGTTACCAGAGCGAAGCACGATGCAAAAAATAAATGTTATGTTTTATCTGGATCAAAAACTTGGATAACCAATGCACCAATCGCTGACATTATTATCGTTTGGGCAAAAGACGACGAAGGTAAAGTACGCGGTTTTATAGTCGAACGAGCCGAGGTAAAGAAAGGTTTAGATACGCCCAAAATCAATGGGAAATTTTCTTTGCGGGCATCGGCAACCGGAATGATATTGCTCGACGAAGTGGTTATACCGGAGGAGAATTTATTGCCGAATGTAGTGGGCCTGAAGGGGCCGTTCGGATGTCTCAATAACGCGCGATACGGTATCGCCTGGGGCGCTCTCGGATCCGCAGAGACGTGTTTGCGAATAGCCCGGCAGTACACATTAGATAGGAAACAATTCGGGCGGCCTTTAGCTTCGAATCAACTTGTACAGAAGAAGCTGGCTGATATGCTGACGGAAATAAATATTGGCTACCAGGCCTGTTTGCGTGTTGGACGTTTAAAAGACGAAGATAAAGCAGCTCCAGAGATGATATCTTTAATTAAGAGAAATAGTTGCGGTAAAGCATTAGAGATTGCCAGAGTAGCAAGAGACATGCTTGGAGGTAATGGGATTTCTGACGAGTATCACATCATACGGCATGTTATGAATTTAGAAGCAGTCAACACTTATGAAGGAACGCACGATATTCATGCGCTTATATTAGGAAGGGCTATTACTGGACTACAAGCTTTTTCTTAA
Protein
MVSLTSKYILSTICKSNSIRALSTTYSRNAKVTFDWVDPFNLDGQLHDDEKAVRDSFRAYCNEKLLPRVVEANRNEVFHREIYNELGELGALGCTIKGYGCAGVSYVTYGLITRELDGVDSSYRSAMSVQSSLAMGSIYMYGTEDQKQKYLPRMATGELIGCFGLTEPNFGSDAGGLVTRAKHDAKNKCYVLSGSKTWITNAPIADIIIVWAKDDEGKVRGFIVERAEVKKGLDTPKINGKFSLRASATGMILLDEVVIPEENLLPNVVGLKGPFGCLNNARYGIAWGALGSAETCLRIARQYTLDRKQFGRPLASNQLVQKKLADMLTEINIGYQACLRVGRLKDEDKAAPEMISLIKRNSCGKALEIARVARDMLGGNGISDEYHIIRHVMNLEAVNTYEGTHDIHALILGRAITGLQAFS
Summary
Cofactor
FAD
Similarity
Belongs to the acyl-CoA dehydrogenase family.
Uniprot
H9J7F2
A0A2A4K1F0
A0A0L7LVE2
A0A194PTJ7
A0A194QSE4
A0A2H1W599
+ More
A0A2M4AET2 U5ER60 A0A182VJL4 A0A1L8DYT9 A0A2M4AF98 A0A2M4AF03 A0A1L8DYW0 A0A0K8TTA5 A0A2M4APA2 A0A2M4AQC0 A0A2M4AGF4 A0A182X167 A0A084VNK7 A0A182L058 A0A1S4GZ75 A0A2C9GS64 W5JFM1 A0A182UAS3 A0A336LV22 A0A336LSF1 Q16IJ3 Q7Q4N7 A0A2M4BP43 A0A1I8JUK1 A0A023ET31 Q16FF1 A0A182VTR8 A0A1I8PUF8 A0A182Y8F4 A0A3B4EX02 T1PD08 A0A1I8MZI9 A0A0L0CQ94 A0A182MD77 A0A3Q2X1Z4 A0A3Q4GSS3 A0A3B3ZAC2 A0A1L8EFS4 A0A1L8EFV8 B0W011 A0A3B4EZR8 A0A3Q2X0V4 A0A3P9AZA2 I3JMS0 A0A3Q3LJN6 A0A1Q3FG09 A0A3Q4H8T9 A0A3P8WD50 A0A3Q0SND2 A0A3P8NCR4 A0A3Q3LJH9 A0A3Q2QGG5 A0A3Q3FNH6 A0A3Q3ENY6 A0A3P8WFZ9 Q6AZV2 A0A147A4A0 A0A3B3X8N9 A4QND3 A0A3Q2DWH7 A0A3Q1IEI1 Q28CC9 F7GD32 A0A3Q3ARZ4 K7G0W0 A0A3Q1IAF4 A0A3B3YMI9 A0A3B3CCZ9 A0A3Q1AWP3 A0A3P8TNN3 A0A2I4B982 A0A3P8TPG0 A0A3B3VV04 A0A3P9LK00 A0A3P9I214 A0A3Q2YD28 A0A3B5QRW4 A0A1J1IXB5 A0A3P9MQJ1 A0A067RA19 A0A3Q1EVJ4 A0A0S7FTW7 A0A3Q3LNH4 A0A0P7UKI8 A0A3B3VV09 H2MPA7 A0A3B3S0X7 A0A3P9I1S2 A0A3B4BT41 A0A3Q2Z9K8 A0A3B3S0Z0 A0A315UVJ2 M4AB06 A0A3P9PE78
A0A2M4AET2 U5ER60 A0A182VJL4 A0A1L8DYT9 A0A2M4AF98 A0A2M4AF03 A0A1L8DYW0 A0A0K8TTA5 A0A2M4APA2 A0A2M4AQC0 A0A2M4AGF4 A0A182X167 A0A084VNK7 A0A182L058 A0A1S4GZ75 A0A2C9GS64 W5JFM1 A0A182UAS3 A0A336LV22 A0A336LSF1 Q16IJ3 Q7Q4N7 A0A2M4BP43 A0A1I8JUK1 A0A023ET31 Q16FF1 A0A182VTR8 A0A1I8PUF8 A0A182Y8F4 A0A3B4EX02 T1PD08 A0A1I8MZI9 A0A0L0CQ94 A0A182MD77 A0A3Q2X1Z4 A0A3Q4GSS3 A0A3B3ZAC2 A0A1L8EFS4 A0A1L8EFV8 B0W011 A0A3B4EZR8 A0A3Q2X0V4 A0A3P9AZA2 I3JMS0 A0A3Q3LJN6 A0A1Q3FG09 A0A3Q4H8T9 A0A3P8WD50 A0A3Q0SND2 A0A3P8NCR4 A0A3Q3LJH9 A0A3Q2QGG5 A0A3Q3FNH6 A0A3Q3ENY6 A0A3P8WFZ9 Q6AZV2 A0A147A4A0 A0A3B3X8N9 A4QND3 A0A3Q2DWH7 A0A3Q1IEI1 Q28CC9 F7GD32 A0A3Q3ARZ4 K7G0W0 A0A3Q1IAF4 A0A3B3YMI9 A0A3B3CCZ9 A0A3Q1AWP3 A0A3P8TNN3 A0A2I4B982 A0A3P8TPG0 A0A3B3VV04 A0A3P9LK00 A0A3P9I214 A0A3Q2YD28 A0A3B5QRW4 A0A1J1IXB5 A0A3P9MQJ1 A0A067RA19 A0A3Q1EVJ4 A0A0S7FTW7 A0A3Q3LNH4 A0A0P7UKI8 A0A3B3VV09 H2MPA7 A0A3B3S0X7 A0A3P9I1S2 A0A3B4BT41 A0A3Q2Z9K8 A0A3B3S0Z0 A0A315UVJ2 M4AB06 A0A3P9PE78
Pubmed
EMBL
BABH01013714
NWSH01000322
PCG77490.1
PCG77491.1
JTDY01000006
KOB79493.1
+ More
KQ459599 KPI94460.1 KQ461154 KPJ08407.1 ODYU01006427 SOQ48260.1 GGFK01005980 MBW39301.1 GANO01002951 JAB56920.1 GFDF01002451 JAV11633.1 GGFK01006096 MBW39417.1 GGFK01006022 MBW39343.1 GFDF01002424 JAV11660.1 GDAI01000220 JAI17383.1 GGFK01009278 MBW42599.1 GGFK01009591 MBW42912.1 GGFK01006549 MBW39870.1 ATLV01014759 KE524984 KFB39551.1 AAAB01008964 APCN01000749 ADMH02001646 ETN61609.1 UFQT01000144 SSX20851.1 SSX20850.1 CH478073 EAT34089.1 EAA12875.3 GGFJ01005641 MBW54782.1 JXUM01070964 GAPW01001637 KQ562637 JAC11961.1 KXJ75440.1 CH478458 EAT32958.1 KA646554 AFP61183.1 JRES01000080 KNC34357.1 AXCM01000962 GFDG01001295 JAV17504.1 GFDG01001294 JAV17505.1 DS231816 EDS37583.1 AERX01009026 GFDL01008570 JAV26475.1 BC077194 CM004470 AAH77194.1 OCT90445.1 GCES01013236 JAR73087.1 BC135458 AAI35459.1 AAMC01089019 AAMC01089020 CR942325 CR761575 CAJ82579.1 CAJ83975.1 AGCU01011279 AGCU01011280 CVRI01000059 CRL03201.1 KK852597 KDR20565.1 GBYX01462812 JAO18782.1 JARO02003472 KPP70418.1 NHOQ01002686 PWA15479.1
KQ459599 KPI94460.1 KQ461154 KPJ08407.1 ODYU01006427 SOQ48260.1 GGFK01005980 MBW39301.1 GANO01002951 JAB56920.1 GFDF01002451 JAV11633.1 GGFK01006096 MBW39417.1 GGFK01006022 MBW39343.1 GFDF01002424 JAV11660.1 GDAI01000220 JAI17383.1 GGFK01009278 MBW42599.1 GGFK01009591 MBW42912.1 GGFK01006549 MBW39870.1 ATLV01014759 KE524984 KFB39551.1 AAAB01008964 APCN01000749 ADMH02001646 ETN61609.1 UFQT01000144 SSX20851.1 SSX20850.1 CH478073 EAT34089.1 EAA12875.3 GGFJ01005641 MBW54782.1 JXUM01070964 GAPW01001637 KQ562637 JAC11961.1 KXJ75440.1 CH478458 EAT32958.1 KA646554 AFP61183.1 JRES01000080 KNC34357.1 AXCM01000962 GFDG01001295 JAV17504.1 GFDG01001294 JAV17505.1 DS231816 EDS37583.1 AERX01009026 GFDL01008570 JAV26475.1 BC077194 CM004470 AAH77194.1 OCT90445.1 GCES01013236 JAR73087.1 BC135458 AAI35459.1 AAMC01089019 AAMC01089020 CR942325 CR761575 CAJ82579.1 CAJ83975.1 AGCU01011279 AGCU01011280 CVRI01000059 CRL03201.1 KK852597 KDR20565.1 GBYX01462812 JAO18782.1 JARO02003472 KPP70418.1 NHOQ01002686 PWA15479.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000075903
+ More
UP000076407 UP000030765 UP000075882 UP000075840 UP000000673 UP000075902 UP000008820 UP000007062 UP000075900 UP000069940 UP000249989 UP000075920 UP000095300 UP000076408 UP000261460 UP000095301 UP000037069 UP000075883 UP000264840 UP000261580 UP000261520 UP000002320 UP000265160 UP000005207 UP000261660 UP000265120 UP000261340 UP000265100 UP000265000 UP000264800 UP000186698 UP000261480 UP000265020 UP000265040 UP000008143 UP000002279 UP000007267 UP000261560 UP000257160 UP000265080 UP000192220 UP000261500 UP000265180 UP000265200 UP000264820 UP000002852 UP000183832 UP000027135 UP000257200 UP000261640 UP000034805 UP000001038 UP000261540 UP000261440 UP000242638
UP000076407 UP000030765 UP000075882 UP000075840 UP000000673 UP000075902 UP000008820 UP000007062 UP000075900 UP000069940 UP000249989 UP000075920 UP000095300 UP000076408 UP000261460 UP000095301 UP000037069 UP000075883 UP000264840 UP000261580 UP000261520 UP000002320 UP000265160 UP000005207 UP000261660 UP000265120 UP000261340 UP000265100 UP000265000 UP000264800 UP000186698 UP000261480 UP000265020 UP000265040 UP000008143 UP000002279 UP000007267 UP000261560 UP000257160 UP000265080 UP000192220 UP000261500 UP000265180 UP000265200 UP000264820 UP000002852 UP000183832 UP000027135 UP000257200 UP000261640 UP000034805 UP000001038 UP000261540 UP000261440 UP000242638
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J7F2
A0A2A4K1F0
A0A0L7LVE2
A0A194PTJ7
A0A194QSE4
A0A2H1W599
+ More
A0A2M4AET2 U5ER60 A0A182VJL4 A0A1L8DYT9 A0A2M4AF98 A0A2M4AF03 A0A1L8DYW0 A0A0K8TTA5 A0A2M4APA2 A0A2M4AQC0 A0A2M4AGF4 A0A182X167 A0A084VNK7 A0A182L058 A0A1S4GZ75 A0A2C9GS64 W5JFM1 A0A182UAS3 A0A336LV22 A0A336LSF1 Q16IJ3 Q7Q4N7 A0A2M4BP43 A0A1I8JUK1 A0A023ET31 Q16FF1 A0A182VTR8 A0A1I8PUF8 A0A182Y8F4 A0A3B4EX02 T1PD08 A0A1I8MZI9 A0A0L0CQ94 A0A182MD77 A0A3Q2X1Z4 A0A3Q4GSS3 A0A3B3ZAC2 A0A1L8EFS4 A0A1L8EFV8 B0W011 A0A3B4EZR8 A0A3Q2X0V4 A0A3P9AZA2 I3JMS0 A0A3Q3LJN6 A0A1Q3FG09 A0A3Q4H8T9 A0A3P8WD50 A0A3Q0SND2 A0A3P8NCR4 A0A3Q3LJH9 A0A3Q2QGG5 A0A3Q3FNH6 A0A3Q3ENY6 A0A3P8WFZ9 Q6AZV2 A0A147A4A0 A0A3B3X8N9 A4QND3 A0A3Q2DWH7 A0A3Q1IEI1 Q28CC9 F7GD32 A0A3Q3ARZ4 K7G0W0 A0A3Q1IAF4 A0A3B3YMI9 A0A3B3CCZ9 A0A3Q1AWP3 A0A3P8TNN3 A0A2I4B982 A0A3P8TPG0 A0A3B3VV04 A0A3P9LK00 A0A3P9I214 A0A3Q2YD28 A0A3B5QRW4 A0A1J1IXB5 A0A3P9MQJ1 A0A067RA19 A0A3Q1EVJ4 A0A0S7FTW7 A0A3Q3LNH4 A0A0P7UKI8 A0A3B3VV09 H2MPA7 A0A3B3S0X7 A0A3P9I1S2 A0A3B4BT41 A0A3Q2Z9K8 A0A3B3S0Z0 A0A315UVJ2 M4AB06 A0A3P9PE78
A0A2M4AET2 U5ER60 A0A182VJL4 A0A1L8DYT9 A0A2M4AF98 A0A2M4AF03 A0A1L8DYW0 A0A0K8TTA5 A0A2M4APA2 A0A2M4AQC0 A0A2M4AGF4 A0A182X167 A0A084VNK7 A0A182L058 A0A1S4GZ75 A0A2C9GS64 W5JFM1 A0A182UAS3 A0A336LV22 A0A336LSF1 Q16IJ3 Q7Q4N7 A0A2M4BP43 A0A1I8JUK1 A0A023ET31 Q16FF1 A0A182VTR8 A0A1I8PUF8 A0A182Y8F4 A0A3B4EX02 T1PD08 A0A1I8MZI9 A0A0L0CQ94 A0A182MD77 A0A3Q2X1Z4 A0A3Q4GSS3 A0A3B3ZAC2 A0A1L8EFS4 A0A1L8EFV8 B0W011 A0A3B4EZR8 A0A3Q2X0V4 A0A3P9AZA2 I3JMS0 A0A3Q3LJN6 A0A1Q3FG09 A0A3Q4H8T9 A0A3P8WD50 A0A3Q0SND2 A0A3P8NCR4 A0A3Q3LJH9 A0A3Q2QGG5 A0A3Q3FNH6 A0A3Q3ENY6 A0A3P8WFZ9 Q6AZV2 A0A147A4A0 A0A3B3X8N9 A4QND3 A0A3Q2DWH7 A0A3Q1IEI1 Q28CC9 F7GD32 A0A3Q3ARZ4 K7G0W0 A0A3Q1IAF4 A0A3B3YMI9 A0A3B3CCZ9 A0A3Q1AWP3 A0A3P8TNN3 A0A2I4B982 A0A3P8TPG0 A0A3B3VV04 A0A3P9LK00 A0A3P9I214 A0A3Q2YD28 A0A3B5QRW4 A0A1J1IXB5 A0A3P9MQJ1 A0A067RA19 A0A3Q1EVJ4 A0A0S7FTW7 A0A3Q3LNH4 A0A0P7UKI8 A0A3B3VV09 H2MPA7 A0A3B3S0X7 A0A3P9I1S2 A0A3B4BT41 A0A3Q2Z9K8 A0A3B3S0Z0 A0A315UVJ2 M4AB06 A0A3P9PE78
PDB
2R0N
E-value=5.54556e-170,
Score=1534
Ontologies
PATHWAY
GO
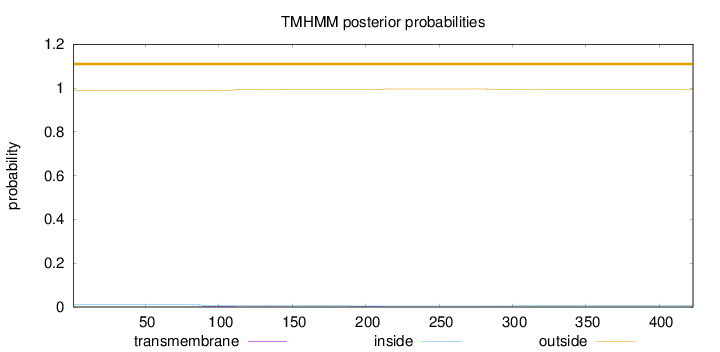
Topology
Length:
423
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18126
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00997
outside
1 - 423
Population Genetic Test Statistics
Pi
18.143813
Theta
13.868107
Tajima's D
0.870694
CLR
4.683808
CSRT
0.629968501574921
Interpretation
Uncertain
