Gene
KWMTBOMO04488
Pre Gene Modal
BGIBMGA005443
Annotation
UDP-glycosyltransferase_UGT39B1_precursor_[Bombyx_mori]
Full name
UDP-glucuronosyltransferase
Location in the cell
PlasmaMembrane Reliability : 3.383
Sequence
CDS
ATGTTCAACAGTTTCATATTTTTGTTTGTTGTAGTGACGGGCTTATGTGAGTCTGCCAATATTTTGTATGTGATGCCTTTTACTTCTAAATCTCACCATATTATGTTGAAGCCGATTGGATTAGAATTAGCTAGAAGGGGTCACAATGTGACCGTCATCACTGGGTTTCGAGATAAGAATGCTCCTGCAAATTACCGTCAGATACAAGTTGATCAGAAAGAAATATGGGACGTCATAGGAACCAAAAGACCGAACGTTTTCGACATGGTCGGGGTATCAACTGAAGAATTTCACAATATGATTCTCTGGAGAGGGGGACTAGGATTCACAAATCTAACTTTAAATTCAGCTGAAGTGAAGAGTTTTTTAGCTGAAGACAACAAATTCGATTTAGTCATTTGCGAGCAGTTTTTCCAAGAAGCGATGAACATATTAGCACATAAATACAAAGCTCCTTTAGTTCTAGTTACCACATTCGGAAACTGTATGAGACATAACATCATGATAAGAAACCCATTGCAGCTAGCCACTGTGATCAGTGAATTTTTGGAAGTGAGGAACCCGACCAGTTTCTTCGCAAGGTTAAGAAATGTATACTTCACCGTTTACGAATACGTTTGGTGGCGATATTGGTACTTAGAAGAACAAGAGAAATTGGTCAAGAAGTACATTCCAAATTTAGAAGAACCAGTGCCCACTTTGCTTGAGATGCAAAAGAATGCCTCCCTGATACTAATTAACGGACATTTTAGTTTTGATACTCCAGCTGCGTATCTTCCCAATATTATTGAAATTGGCGGAGTGCATCTTTCCAAGAGTGACACTAAGCTGCCCGCGGATTTGCAAAACATTTTAGACGAAGCAAAGCATGGAGTCATTTACATTAACTTTGGATCGAACGTGCGTAGTGCTGAACTACCGTTAGAAAAAAGAAATGTATTTTTGAATGTAATTAAGAAACTGAAGCAGACTGTGGTGTGGAAATGGGAAGATGACAGTCTAGATAAAATGGATAATTTAGTCGTGAGAAAATGGCTACCACAAAAAGAAATATTATCCCACCCAAATATCAAAGTATTCATATCGCATGGTGGATTGATCGGAACACAGGAAGCCATATTCCACGGCGTGCCTATAATTGGAGTTCCTATTTACGCTGACCAATACAATAATTTATTACAAGCAGAAGAAATTGGATTTGGGAAAATACTCGAATTCAAAGATATCAGAGAACAAAACTTAGATAATTACTTGAGAGAGCTTCTTACAAACAATACTTACAGAGACAAAGCTAAAGAGATGTCTATTAGGTTTAAAGATAGACCGACGACTGCTTTAGATACAGCTATGTATTGGATTGAATATATTATAAGGCATAATGGGGCTAGTTTTATGAAAAATCCAGCCAGAAAATTGCACTGGATCCAATACGCAATGCTAGATGTTTATGGTTTTATATTAGCAGTAGTGTTAACTATTTTTTACACTATTTACAAACTAAGCAGTTTCATATTGCATAAATTGAAAGCTCCTGAGAGACTTATTAGAAAAAAGTTTGATTGA
Protein
MFNSFIFLFVVVTGLCESANILYVMPFTSKSHHIMLKPIGLELARRGHNVTVITGFRDKNAPANYRQIQVDQKEIWDVIGTKRPNVFDMVGVSTEEFHNMILWRGGLGFTNLTLNSAEVKSFLAEDNKFDLVICEQFFQEAMNILAHKYKAPLVLVTTFGNCMRHNIMIRNPLQLATVISEFLEVRNPTSFFARLRNVYFTVYEYVWWRYWYLEEQEKLVKKYIPNLEEPVPTLLEMQKNASLILINGHFSFDTPAAYLPNIIEIGGVHLSKSDTKLPADLQNILDEAKHGVIYINFGSNVRSAELPLEKRNVFLNVIKKLKQTVVWKWEDDSLDKMDNLVVRKWLPQKEILSHPNIKVFISHGGLIGTQEAIFHGVPIIGVPIYADQYNNLLQAEEIGFGKILEFKDIREQNLDNYLRELLTNNTYRDKAKEMSIRFKDRPTTALDTAMYWIEYIIRHNGASFMKNPARKLHWIQYAMLDVYGFILAVVLTIFYTIYKLSSFILHKLKAPERLIRKKFD
Summary
Catalytic Activity
glucuronate acceptor + UDP-alpha-D-glucuronate = acceptor beta-D-glucuronoside + H(+) + UDP
Similarity
Belongs to the UDP-glycosyltransferase family.
Belongs to the ubiquitin-conjugating enzyme family.
Belongs to the ubiquitin-conjugating enzyme family.
Feature
chain UDP-glucuronosyltransferase
Uniprot
G9LPT8
H9J7F1
A0A2H1W5A1
A0A191T1L9
G9LPP9
A0A3S2N8S7
+ More
A0A2A4K0B3 A0A212FL56 A0A2H4WB64 A0A1L8D6G2 A0A194PNY6 A0A212EHV3 G9LPT9 A0A0L7LWH9 A0A194QSA9 D2JLL0 H9J7F0 A0A2H4WB65 A0A2M4BJX2 A0A2M4BJY0 A0A2M4BJY3 A0A182IJQ8 A0A182FDS8 A0A182M8S5 W5JEF5 A0A2M3Z3U6 A0A2M4AGW6 A0A2M4AFV3 A0A2M3Z343 A0A084WK54 A0A182Y289 A0A182WL35 A0A182TC71 A0A2M4AGI4 Q7PTF6 A0A182NJM1 A0A182JUT3 A0A182I6Z1 A0A182R334 A0A182QE42 A0A182L3C8 A0A182WWQ5 A0A182PQN3 A0A182VHN2 A0A182TJ21 A0A1Y9HEP2 A0A1Q3FRS4 A0A1Q3FZT9 B0W9Y5 A0A0M4EDN6 A0A182R101 A0A182JQE6 A0A182XYP3 A0A2J7Q382 B4KPM7 A0A0M3QVA8 Q17GE2 Q17GE3 V5I8H8 U5EXU4 B4LYF5 A0A182WJE5 A0A182M4J5 B4J7P4 A0A2M4CI25 A0A2M3Z5E9 A0A1S4GSA2 A0A2J7Q396 B4KPM8 A0A182NI27 B4MRB3 Q1HRH8 A0A1S4FQK6 A0A2M4BK13 A0A2M3Z5C0 A0A182FDF0 A0A2M4CHZ1 A0A182PQM1 A0A182IIM5 A0A2M4BJZ5 A0A1Y1NF76 A0A023EW79 A0A2M3ZF25 A0A067RFX2 B3MBV3 A0A1A9ZAS7 A0A067R1S8 A0A2J7Q393 A0A2M4AFS8 A0A2J7Q399 A0A1B0DAR4 A0A2M4BKA0 A0A2M4BK77 A0A1Q3FZT2 A0A0K8UF88 A0A1W4UAS2 A0A1I8JUM7 Q7Q5T0 B4GA69 A0A2M4AFT4
A0A2A4K0B3 A0A212FL56 A0A2H4WB64 A0A1L8D6G2 A0A194PNY6 A0A212EHV3 G9LPT9 A0A0L7LWH9 A0A194QSA9 D2JLL0 H9J7F0 A0A2H4WB65 A0A2M4BJX2 A0A2M4BJY0 A0A2M4BJY3 A0A182IJQ8 A0A182FDS8 A0A182M8S5 W5JEF5 A0A2M3Z3U6 A0A2M4AGW6 A0A2M4AFV3 A0A2M3Z343 A0A084WK54 A0A182Y289 A0A182WL35 A0A182TC71 A0A2M4AGI4 Q7PTF6 A0A182NJM1 A0A182JUT3 A0A182I6Z1 A0A182R334 A0A182QE42 A0A182L3C8 A0A182WWQ5 A0A182PQN3 A0A182VHN2 A0A182TJ21 A0A1Y9HEP2 A0A1Q3FRS4 A0A1Q3FZT9 B0W9Y5 A0A0M4EDN6 A0A182R101 A0A182JQE6 A0A182XYP3 A0A2J7Q382 B4KPM7 A0A0M3QVA8 Q17GE2 Q17GE3 V5I8H8 U5EXU4 B4LYF5 A0A182WJE5 A0A182M4J5 B4J7P4 A0A2M4CI25 A0A2M3Z5E9 A0A1S4GSA2 A0A2J7Q396 B4KPM8 A0A182NI27 B4MRB3 Q1HRH8 A0A1S4FQK6 A0A2M4BK13 A0A2M3Z5C0 A0A182FDF0 A0A2M4CHZ1 A0A182PQM1 A0A182IIM5 A0A2M4BJZ5 A0A1Y1NF76 A0A023EW79 A0A2M3ZF25 A0A067RFX2 B3MBV3 A0A1A9ZAS7 A0A067R1S8 A0A2J7Q393 A0A2M4AFS8 A0A2J7Q399 A0A1B0DAR4 A0A2M4BKA0 A0A2M4BK77 A0A1Q3FZT2 A0A0K8UF88 A0A1W4UAS2 A0A1I8JUM7 Q7Q5T0 B4GA69 A0A2M4AFT4
EC Number
2.4.1.17
Pubmed
EMBL
JQ070245
AEW43161.1
BABH01013714
ODYU01006427
SOQ48259.1
KU680293
+ More
ANI21999.1 JQ070206 AEW43122.1 RSAL01000196 RVE44553.1 NWSH01000322 PCG77489.1 AGBW02007908 OWR54430.1 KY202931 AUC64275.1 GEYN01000086 JAV02043.1 KQ459599 KPI94459.1 AGBW02014796 OWR41057.1 JQ070246 AEW43162.1 JTDY01000006 KOB79496.1 KQ461154 KPJ08408.1 GQ915326 ACZ97420.2 KY202935 AUC64279.1 GGFJ01004214 MBW53355.1 GGFJ01004173 MBW53314.1 GGFJ01004216 MBW53357.1 AXCM01005982 ADMH02001746 ETN61209.1 GGFM01002441 MBW23192.1 GGFK01006656 MBW39977.1 GGFK01006354 MBW39675.1 GGFM01002198 MBW22949.1 ATLV01024094 KE525349 KFB50598.1 GGFK01006565 MBW39886.1 AAAB01008807 EAA03993.4 APCN01003712 AXCN02000438 GFDL01004778 JAV30267.1 GFDL01001967 JAV33078.1 DS231867 EDS40643.1 CP012524 ALC42052.1 AXCN02000388 NEVH01019069 PNF23045.1 CH933808 EDW09137.1 ALC42053.1 CH477262 EAT45644.1 EAT45643.1 GALX01004660 JAB63806.1 GANO01000777 JAB59094.1 CH940650 EDW66951.1 AXCM01000608 CH916367 EDW02192.1 GGFL01000775 MBW64953.1 GGFM01002996 MBW23747.1 AAAB01008960 PNF23054.1 EDW09138.2 CH963850 EDW74652.1 DQ440116 ABF18149.1 GGFJ01004254 MBW53395.1 GGFM01002966 MBW23717.1 GGFL01000776 MBW64954.1 APCN01000032 GGFJ01004255 MBW53396.1 GEZM01007277 JAV95265.1 GAPW01000969 JAC12629.1 GGFM01006340 MBW27091.1 KK852488 KDR22771.1 CH902619 EDV36124.2 KK853091 KDR11536.1 PNF23046.1 GGFK01006318 MBW39639.1 PNF23049.1 AJVK01013380 GGFJ01004240 MBW53381.1 GGFJ01004316 MBW53457.1 GFDL01001981 JAV33064.1 GDHF01026977 JAI25337.1 EAA11593.4 CH479181 EDW31821.1 GGFK01006329 MBW39650.1
ANI21999.1 JQ070206 AEW43122.1 RSAL01000196 RVE44553.1 NWSH01000322 PCG77489.1 AGBW02007908 OWR54430.1 KY202931 AUC64275.1 GEYN01000086 JAV02043.1 KQ459599 KPI94459.1 AGBW02014796 OWR41057.1 JQ070246 AEW43162.1 JTDY01000006 KOB79496.1 KQ461154 KPJ08408.1 GQ915326 ACZ97420.2 KY202935 AUC64279.1 GGFJ01004214 MBW53355.1 GGFJ01004173 MBW53314.1 GGFJ01004216 MBW53357.1 AXCM01005982 ADMH02001746 ETN61209.1 GGFM01002441 MBW23192.1 GGFK01006656 MBW39977.1 GGFK01006354 MBW39675.1 GGFM01002198 MBW22949.1 ATLV01024094 KE525349 KFB50598.1 GGFK01006565 MBW39886.1 AAAB01008807 EAA03993.4 APCN01003712 AXCN02000438 GFDL01004778 JAV30267.1 GFDL01001967 JAV33078.1 DS231867 EDS40643.1 CP012524 ALC42052.1 AXCN02000388 NEVH01019069 PNF23045.1 CH933808 EDW09137.1 ALC42053.1 CH477262 EAT45644.1 EAT45643.1 GALX01004660 JAB63806.1 GANO01000777 JAB59094.1 CH940650 EDW66951.1 AXCM01000608 CH916367 EDW02192.1 GGFL01000775 MBW64953.1 GGFM01002996 MBW23747.1 AAAB01008960 PNF23054.1 EDW09138.2 CH963850 EDW74652.1 DQ440116 ABF18149.1 GGFJ01004254 MBW53395.1 GGFM01002966 MBW23717.1 GGFL01000776 MBW64954.1 APCN01000032 GGFJ01004255 MBW53396.1 GEZM01007277 JAV95265.1 GAPW01000969 JAC12629.1 GGFM01006340 MBW27091.1 KK852488 KDR22771.1 CH902619 EDV36124.2 KK853091 KDR11536.1 PNF23046.1 GGFK01006318 MBW39639.1 PNF23049.1 AJVK01013380 GGFJ01004240 MBW53381.1 GGFJ01004316 MBW53457.1 GFDL01001981 JAV33064.1 GDHF01026977 JAI25337.1 EAA11593.4 CH479181 EDW31821.1 GGFK01006329 MBW39650.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053268
UP000037510
+ More
UP000053240 UP000075880 UP000069272 UP000075883 UP000000673 UP000030765 UP000076408 UP000075920 UP000075901 UP000007062 UP000075884 UP000075881 UP000075840 UP000075900 UP000075886 UP000075882 UP000076407 UP000075885 UP000075903 UP000075902 UP000002320 UP000092553 UP000235965 UP000009192 UP000008820 UP000008792 UP000001070 UP000007798 UP000027135 UP000007801 UP000092445 UP000092462 UP000192221 UP000008744
UP000053240 UP000075880 UP000069272 UP000075883 UP000000673 UP000030765 UP000076408 UP000075920 UP000075901 UP000007062 UP000075884 UP000075881 UP000075840 UP000075900 UP000075886 UP000075882 UP000076407 UP000075885 UP000075903 UP000075902 UP000002320 UP000092553 UP000235965 UP000009192 UP000008820 UP000008792 UP000001070 UP000007798 UP000027135 UP000007801 UP000092445 UP000092462 UP000192221 UP000008744
Interpro
SUPFAM
SSF54495
SSF54495
Gene 3D
CDD
ProteinModelPortal
G9LPT8
H9J7F1
A0A2H1W5A1
A0A191T1L9
G9LPP9
A0A3S2N8S7
+ More
A0A2A4K0B3 A0A212FL56 A0A2H4WB64 A0A1L8D6G2 A0A194PNY6 A0A212EHV3 G9LPT9 A0A0L7LWH9 A0A194QSA9 D2JLL0 H9J7F0 A0A2H4WB65 A0A2M4BJX2 A0A2M4BJY0 A0A2M4BJY3 A0A182IJQ8 A0A182FDS8 A0A182M8S5 W5JEF5 A0A2M3Z3U6 A0A2M4AGW6 A0A2M4AFV3 A0A2M3Z343 A0A084WK54 A0A182Y289 A0A182WL35 A0A182TC71 A0A2M4AGI4 Q7PTF6 A0A182NJM1 A0A182JUT3 A0A182I6Z1 A0A182R334 A0A182QE42 A0A182L3C8 A0A182WWQ5 A0A182PQN3 A0A182VHN2 A0A182TJ21 A0A1Y9HEP2 A0A1Q3FRS4 A0A1Q3FZT9 B0W9Y5 A0A0M4EDN6 A0A182R101 A0A182JQE6 A0A182XYP3 A0A2J7Q382 B4KPM7 A0A0M3QVA8 Q17GE2 Q17GE3 V5I8H8 U5EXU4 B4LYF5 A0A182WJE5 A0A182M4J5 B4J7P4 A0A2M4CI25 A0A2M3Z5E9 A0A1S4GSA2 A0A2J7Q396 B4KPM8 A0A182NI27 B4MRB3 Q1HRH8 A0A1S4FQK6 A0A2M4BK13 A0A2M3Z5C0 A0A182FDF0 A0A2M4CHZ1 A0A182PQM1 A0A182IIM5 A0A2M4BJZ5 A0A1Y1NF76 A0A023EW79 A0A2M3ZF25 A0A067RFX2 B3MBV3 A0A1A9ZAS7 A0A067R1S8 A0A2J7Q393 A0A2M4AFS8 A0A2J7Q399 A0A1B0DAR4 A0A2M4BKA0 A0A2M4BK77 A0A1Q3FZT2 A0A0K8UF88 A0A1W4UAS2 A0A1I8JUM7 Q7Q5T0 B4GA69 A0A2M4AFT4
A0A2A4K0B3 A0A212FL56 A0A2H4WB64 A0A1L8D6G2 A0A194PNY6 A0A212EHV3 G9LPT9 A0A0L7LWH9 A0A194QSA9 D2JLL0 H9J7F0 A0A2H4WB65 A0A2M4BJX2 A0A2M4BJY0 A0A2M4BJY3 A0A182IJQ8 A0A182FDS8 A0A182M8S5 W5JEF5 A0A2M3Z3U6 A0A2M4AGW6 A0A2M4AFV3 A0A2M3Z343 A0A084WK54 A0A182Y289 A0A182WL35 A0A182TC71 A0A2M4AGI4 Q7PTF6 A0A182NJM1 A0A182JUT3 A0A182I6Z1 A0A182R334 A0A182QE42 A0A182L3C8 A0A182WWQ5 A0A182PQN3 A0A182VHN2 A0A182TJ21 A0A1Y9HEP2 A0A1Q3FRS4 A0A1Q3FZT9 B0W9Y5 A0A0M4EDN6 A0A182R101 A0A182JQE6 A0A182XYP3 A0A2J7Q382 B4KPM7 A0A0M3QVA8 Q17GE2 Q17GE3 V5I8H8 U5EXU4 B4LYF5 A0A182WJE5 A0A182M4J5 B4J7P4 A0A2M4CI25 A0A2M3Z5E9 A0A1S4GSA2 A0A2J7Q396 B4KPM8 A0A182NI27 B4MRB3 Q1HRH8 A0A1S4FQK6 A0A2M4BK13 A0A2M3Z5C0 A0A182FDF0 A0A2M4CHZ1 A0A182PQM1 A0A182IIM5 A0A2M4BJZ5 A0A1Y1NF76 A0A023EW79 A0A2M3ZF25 A0A067RFX2 B3MBV3 A0A1A9ZAS7 A0A067R1S8 A0A2J7Q393 A0A2M4AFS8 A0A2J7Q399 A0A1B0DAR4 A0A2M4BKA0 A0A2M4BK77 A0A1Q3FZT2 A0A0K8UF88 A0A1W4UAS2 A0A1I8JUM7 Q7Q5T0 B4GA69 A0A2M4AFT4
PDB
2O6L
E-value=1.87879e-22,
Score=263
Ontologies
PATHWAY
00040
Pentose and glucuronate interconversions - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
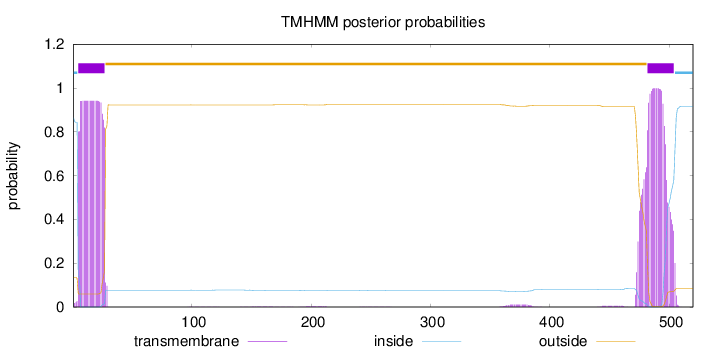
Topology
Subcellular location
Membrane
SignalP
Position: 1 - 18,
Likelihood: 0.969764

Length:
520
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
44.66265
Exp number, first 60 AAs:
21.35964
Total prob of N-in:
0.86424
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 481
TMhelix
482 - 504
inside
505 - 520
Population Genetic Test Statistics
Pi
29.980973
Theta
35.513122
Tajima's D
-0.558114
CLR
1.432673
CSRT
0.225988700564972
Interpretation
Uncertain
