Gene
KWMTBOMO04481
Annotation
PREDICTED:_vitamin_K_epoxide_reductase_complex_subunit_1_[Plutella_xylostella]
Location in the cell
PlasmaMembrane Reliability : 4.321
Sequence
CDS
ATGAAGGTAAGAACATACGACCGTTTGCTAATACTACTTTCGGTTCTCGGAGTTTTACTTTCAACCTATGCACTGTATGTGGAGCTGAAAGTGGAGGCAAATCCCAATTATAAAGCCTTTTGCGACTTTTCCGAGCGAGCAAGTTGCACGAGAGTGCTTACTTCGAAGTATGCCAAAGGTCTTGGAATAATCGACGAAGGATCATATTTACGTCTTCCGAATCCAATTTATGGAACAATTCACTTCTGCATTATGATCATTTTAAGTGCTTACGACAGTTTGGTTACGACAAATATTCAATTTTCCCTCGCTGGAATATCTTCACTGCTGTGCTGCTATTTAGCGTATAATCTTATTTACGTTTTAAAAGACATTTGTATAGTTTGTATATCTACTTACTTGATAAATGGAACTATGTTATTTTTGATTTGGAGCAAATTTAAACGTCAAGCAAAGAAAATTGCTTAA
Protein
MKVRTYDRLLILLSVLGVLLSTYALYVELKVEANPNYKAFCDFSERASCTRVLTSKYAKGLGIIDEGSYLRLPNPIYGTIHFCIMIILSAYDSLVTTNIQFSLAGISSLLCCYLAYNLIYVLKDICIVCISTYLINGTMLFLIWSKFKRQAKKIA
Summary
Similarity
Belongs to the cytochrome b5 family.
Uniprot
A0A2A4JAF1
S4PSG6
A0A2H1W9H5
A0A194QSB4
A0A212ER98
A0A1E1WVC0
+ More
A0A1E1W0A6 A0A1E1WUY6 B3RK09 A0A0L7R5L9 A0A1S3GXR5 A0A2R7VWW0 T1GFC2 W4Z817 A0A0M9ABH4 J3JUW5 C3ZV73 F2U6U7 A0A336LL03 A0A0L8GF89 Q16QG8 T1J6K7 A0A1L8DUT8 A0A1Y1M833 Q1HR80 A0A2G8K3X9 E2AVC2 V4C6M3 A7RY76 A0A2P6KSF9 A0A3M7RN27 A0A0P4VYP9 A0A2P8XW72 A0A0V0G774 A0A0P5QJ23 A0A0N8C2N8 A0A0P5QGX7 A0A0P5HM13 A0A0P5RA20 A0A0P5MNN2 A0A0P5S2M6 A0A0P5FQ20 A0A0N8BPN4 A0A0P6HT18 A0A0P5JJK9 A0A0P5NJU5 A0A0P6D3Q1 A0A0P5RVH1 A0A0P5KVK1 A0A0P6EC03 A0A195EEQ8 A0A310SFU5 E9GLS7 A0A0P5LFK4 F4WQY8 K1Q1U5 E0VCW2 A0A0P5GA51 A0A158NTL7 A0A0P5J7G8 A0A0P6E4D8 A0A069DPR8 A0A1D2NJ59 A0A0P5KJ71 A0A0P5IM56 A0A0P5Z6D9 C3XVV5 A0A0P5BUM2 A0A026VZ16 A0A0P5KW66 A0A023GNY9 A0A0P5KZR6 G3MR58 A0A0P5AHE6 T1KBS0 A0A0P5YG89 A0A0P5E293 A0A0P5YUI7 A0A224XV21 A0A2B4S7Y4 A0A0P5X1W3 A0A0P5W592 A0A0P5Z889 A0A0P5B2Q3 A0A0N8D179 A0A0P5VW13 A0A3R7LZU9 A0A0P5SX60 A0A0N7ZR73 A0A0P5EC66 A0A0P4YNZ5 A0A0P5SW50 A0A0P6D029 A0A1B0EUN5 A0A0P4Z434 A0A0P5AC32 A0A0N8D8J1 A0A0P6BWX9 A0A2H1BVZ3
A0A1E1W0A6 A0A1E1WUY6 B3RK09 A0A0L7R5L9 A0A1S3GXR5 A0A2R7VWW0 T1GFC2 W4Z817 A0A0M9ABH4 J3JUW5 C3ZV73 F2U6U7 A0A336LL03 A0A0L8GF89 Q16QG8 T1J6K7 A0A1L8DUT8 A0A1Y1M833 Q1HR80 A0A2G8K3X9 E2AVC2 V4C6M3 A7RY76 A0A2P6KSF9 A0A3M7RN27 A0A0P4VYP9 A0A2P8XW72 A0A0V0G774 A0A0P5QJ23 A0A0N8C2N8 A0A0P5QGX7 A0A0P5HM13 A0A0P5RA20 A0A0P5MNN2 A0A0P5S2M6 A0A0P5FQ20 A0A0N8BPN4 A0A0P6HT18 A0A0P5JJK9 A0A0P5NJU5 A0A0P6D3Q1 A0A0P5RVH1 A0A0P5KVK1 A0A0P6EC03 A0A195EEQ8 A0A310SFU5 E9GLS7 A0A0P5LFK4 F4WQY8 K1Q1U5 E0VCW2 A0A0P5GA51 A0A158NTL7 A0A0P5J7G8 A0A0P6E4D8 A0A069DPR8 A0A1D2NJ59 A0A0P5KJ71 A0A0P5IM56 A0A0P5Z6D9 C3XVV5 A0A0P5BUM2 A0A026VZ16 A0A0P5KW66 A0A023GNY9 A0A0P5KZR6 G3MR58 A0A0P5AHE6 T1KBS0 A0A0P5YG89 A0A0P5E293 A0A0P5YUI7 A0A224XV21 A0A2B4S7Y4 A0A0P5X1W3 A0A0P5W592 A0A0P5Z889 A0A0P5B2Q3 A0A0N8D179 A0A0P5VW13 A0A3R7LZU9 A0A0P5SX60 A0A0N7ZR73 A0A0P5EC66 A0A0P4YNZ5 A0A0P5SW50 A0A0P6D029 A0A1B0EUN5 A0A0P4Z434 A0A0P5AC32 A0A0N8D8J1 A0A0P6BWX9 A0A2H1BVZ3
Pubmed
EMBL
NWSH01002334
PCG68504.1
GAIX01013823
JAA78737.1
ODYU01007148
SOQ49673.1
+ More
KQ461154 KPJ08413.1 AGBW02013068 OWR44020.1 GDQN01000130 JAT90924.1 GDQN01010690 JAT80364.1 GDQN01000224 JAT90830.1 DS985241 EDV28554.1 KQ414652 KOC66114.1 KK854107 PTY11391.1 CAQQ02392320 AAGJ04061500 KQ435710 KOX79863.1 APGK01056839 BT127030 KB741277 KB632028 AEE61992.1 ENN71153.1 ERL88114.1 GG666687 EEN43588.1 GL832963 EGD83579.1 UFQS01000005 UFQT01000005 SSW96952.1 SSX17339.1 KQ422047 KOF75618.1 CH477748 EAT36646.1 JH431878 GFDF01003892 JAV10192.1 GEZM01041995 GEZM01041994 JAV80076.1 DQ440214 ABF18247.1 MRZV01000910 PIK42708.1 GL443064 EFN62618.1 KB201305 ESO97314.1 DS469552 EDO43588.1 MWRG01005942 PRD29281.1 REGN01003009 RNA24946.1 GDRN01086641 JAI61137.1 PYGN01001255 PSN36255.1 GECL01002512 JAP03612.1 GDIQ01123354 JAL28372.1 GDIQ01120755 JAL30971.1 GDIQ01120754 JAL30972.1 GDIQ01231463 JAK20262.1 GDIQ01123353 JAL28373.1 GDIQ01160780 JAK90945.1 GDIQ01093158 JAL58568.1 GDIQ01259522 JAJ92202.1 GDIQ01157187 JAK94538.1 GDIQ01039280 JAN55457.1 GDIQ01204120 JAK47605.1 GDIQ01141193 JAL10533.1 GDIQ01083365 JAN11372.1 GDIQ01102962 JAL48764.1 GDIQ01186522 JAK65203.1 GDIQ01070774 JAN23963.1 KQ978983 KYN26760.1 KQ764881 OAD54340.1 GL732551 EFX79558.1 GDIQ01170332 JAK81393.1 GL888275 EGI63501.1 JH817326 EKC30387.1 DS235065 EEB11218.1 GDIQ01249204 JAK02521.1 ADTU01026019 GDIQ01204119 JAK47606.1 GDIQ01069125 JAN25612.1 GBGD01003267 JAC85622.1 LJIJ01000026 ODN05294.1 GDIQ01186523 GDIQ01184927 JAK66798.1 GDIQ01210761 JAK40964.1 GDIP01050412 JAM53303.1 GG666469 EEN67898.1 GDIP01183940 JAJ39462.1 KK107549 QOIP01000010 EZA49048.1 RLU17304.1 GDIQ01204121 JAK47604.1 GBBM01000355 JAC35063.1 GDIQ01184926 JAK66799.1 JO844359 AEO35976.1 GDIP01199503 JAJ23899.1 CAEY01001954 GDIP01058318 JAM45397.1 GDIP01147463 JAJ75939.1 GDIP01052922 JAM50793.1 GFTR01002748 JAW13678.1 LSMT01000169 PFX24717.1 GDIP01078814 JAM24901.1 GDIP01091069 JAM12646.1 GDIP01047722 JAM55993.1 GDIP01190604 JAJ32798.1 GDIP01078815 JAM24900.1 GDIP01094458 JAM09257.1 QCYY01003438 ROT63451.1 GDIP01134113 JAL69601.1 GDIP01220553 JAJ02849.1 GDIP01150062 JAJ73340.1 GDIP01225890 JAI97511.1 GDIP01139462 JAL64252.1 GDIQ01084917 JAN09820.1 AJWK01014107 AJWK01014108 GDIP01219696 JAJ03706.1 GDIP01202054 LRGB01000944 JAJ21348.1 KZS14479.1 GDIP01058319 JAM45396.1 GDIP01009378 JAM94337.1 KZ433702 PIS79364.1
KQ461154 KPJ08413.1 AGBW02013068 OWR44020.1 GDQN01000130 JAT90924.1 GDQN01010690 JAT80364.1 GDQN01000224 JAT90830.1 DS985241 EDV28554.1 KQ414652 KOC66114.1 KK854107 PTY11391.1 CAQQ02392320 AAGJ04061500 KQ435710 KOX79863.1 APGK01056839 BT127030 KB741277 KB632028 AEE61992.1 ENN71153.1 ERL88114.1 GG666687 EEN43588.1 GL832963 EGD83579.1 UFQS01000005 UFQT01000005 SSW96952.1 SSX17339.1 KQ422047 KOF75618.1 CH477748 EAT36646.1 JH431878 GFDF01003892 JAV10192.1 GEZM01041995 GEZM01041994 JAV80076.1 DQ440214 ABF18247.1 MRZV01000910 PIK42708.1 GL443064 EFN62618.1 KB201305 ESO97314.1 DS469552 EDO43588.1 MWRG01005942 PRD29281.1 REGN01003009 RNA24946.1 GDRN01086641 JAI61137.1 PYGN01001255 PSN36255.1 GECL01002512 JAP03612.1 GDIQ01123354 JAL28372.1 GDIQ01120755 JAL30971.1 GDIQ01120754 JAL30972.1 GDIQ01231463 JAK20262.1 GDIQ01123353 JAL28373.1 GDIQ01160780 JAK90945.1 GDIQ01093158 JAL58568.1 GDIQ01259522 JAJ92202.1 GDIQ01157187 JAK94538.1 GDIQ01039280 JAN55457.1 GDIQ01204120 JAK47605.1 GDIQ01141193 JAL10533.1 GDIQ01083365 JAN11372.1 GDIQ01102962 JAL48764.1 GDIQ01186522 JAK65203.1 GDIQ01070774 JAN23963.1 KQ978983 KYN26760.1 KQ764881 OAD54340.1 GL732551 EFX79558.1 GDIQ01170332 JAK81393.1 GL888275 EGI63501.1 JH817326 EKC30387.1 DS235065 EEB11218.1 GDIQ01249204 JAK02521.1 ADTU01026019 GDIQ01204119 JAK47606.1 GDIQ01069125 JAN25612.1 GBGD01003267 JAC85622.1 LJIJ01000026 ODN05294.1 GDIQ01186523 GDIQ01184927 JAK66798.1 GDIQ01210761 JAK40964.1 GDIP01050412 JAM53303.1 GG666469 EEN67898.1 GDIP01183940 JAJ39462.1 KK107549 QOIP01000010 EZA49048.1 RLU17304.1 GDIQ01204121 JAK47604.1 GBBM01000355 JAC35063.1 GDIQ01184926 JAK66799.1 JO844359 AEO35976.1 GDIP01199503 JAJ23899.1 CAEY01001954 GDIP01058318 JAM45397.1 GDIP01147463 JAJ75939.1 GDIP01052922 JAM50793.1 GFTR01002748 JAW13678.1 LSMT01000169 PFX24717.1 GDIP01078814 JAM24901.1 GDIP01091069 JAM12646.1 GDIP01047722 JAM55993.1 GDIP01190604 JAJ32798.1 GDIP01078815 JAM24900.1 GDIP01094458 JAM09257.1 QCYY01003438 ROT63451.1 GDIP01134113 JAL69601.1 GDIP01220553 JAJ02849.1 GDIP01150062 JAJ73340.1 GDIP01225890 JAI97511.1 GDIP01139462 JAL64252.1 GDIQ01084917 JAN09820.1 AJWK01014107 AJWK01014108 GDIP01219696 JAJ03706.1 GDIP01202054 LRGB01000944 JAJ21348.1 KZS14479.1 GDIP01058319 JAM45396.1 GDIP01009378 JAM94337.1 KZ433702 PIS79364.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000009022
UP000053825
UP000085678
+ More
UP000015102 UP000007110 UP000053105 UP000019118 UP000030742 UP000001554 UP000007799 UP000053454 UP000008820 UP000230750 UP000000311 UP000030746 UP000001593 UP000276133 UP000245037 UP000078492 UP000000305 UP000007755 UP000005408 UP000009046 UP000005205 UP000094527 UP000053097 UP000279307 UP000015104 UP000225706 UP000283509 UP000092461 UP000076858
UP000015102 UP000007110 UP000053105 UP000019118 UP000030742 UP000001554 UP000007799 UP000053454 UP000008820 UP000230750 UP000000311 UP000030746 UP000001593 UP000276133 UP000245037 UP000078492 UP000000305 UP000007755 UP000005408 UP000009046 UP000005205 UP000094527 UP000053097 UP000279307 UP000015104 UP000225706 UP000283509 UP000092461 UP000076858
Interpro
SUPFAM
SSF55856
SSF55856
Gene 3D
ProteinModelPortal
A0A2A4JAF1
S4PSG6
A0A2H1W9H5
A0A194QSB4
A0A212ER98
A0A1E1WVC0
+ More
A0A1E1W0A6 A0A1E1WUY6 B3RK09 A0A0L7R5L9 A0A1S3GXR5 A0A2R7VWW0 T1GFC2 W4Z817 A0A0M9ABH4 J3JUW5 C3ZV73 F2U6U7 A0A336LL03 A0A0L8GF89 Q16QG8 T1J6K7 A0A1L8DUT8 A0A1Y1M833 Q1HR80 A0A2G8K3X9 E2AVC2 V4C6M3 A7RY76 A0A2P6KSF9 A0A3M7RN27 A0A0P4VYP9 A0A2P8XW72 A0A0V0G774 A0A0P5QJ23 A0A0N8C2N8 A0A0P5QGX7 A0A0P5HM13 A0A0P5RA20 A0A0P5MNN2 A0A0P5S2M6 A0A0P5FQ20 A0A0N8BPN4 A0A0P6HT18 A0A0P5JJK9 A0A0P5NJU5 A0A0P6D3Q1 A0A0P5RVH1 A0A0P5KVK1 A0A0P6EC03 A0A195EEQ8 A0A310SFU5 E9GLS7 A0A0P5LFK4 F4WQY8 K1Q1U5 E0VCW2 A0A0P5GA51 A0A158NTL7 A0A0P5J7G8 A0A0P6E4D8 A0A069DPR8 A0A1D2NJ59 A0A0P5KJ71 A0A0P5IM56 A0A0P5Z6D9 C3XVV5 A0A0P5BUM2 A0A026VZ16 A0A0P5KW66 A0A023GNY9 A0A0P5KZR6 G3MR58 A0A0P5AHE6 T1KBS0 A0A0P5YG89 A0A0P5E293 A0A0P5YUI7 A0A224XV21 A0A2B4S7Y4 A0A0P5X1W3 A0A0P5W592 A0A0P5Z889 A0A0P5B2Q3 A0A0N8D179 A0A0P5VW13 A0A3R7LZU9 A0A0P5SX60 A0A0N7ZR73 A0A0P5EC66 A0A0P4YNZ5 A0A0P5SW50 A0A0P6D029 A0A1B0EUN5 A0A0P4Z434 A0A0P5AC32 A0A0N8D8J1 A0A0P6BWX9 A0A2H1BVZ3
A0A1E1W0A6 A0A1E1WUY6 B3RK09 A0A0L7R5L9 A0A1S3GXR5 A0A2R7VWW0 T1GFC2 W4Z817 A0A0M9ABH4 J3JUW5 C3ZV73 F2U6U7 A0A336LL03 A0A0L8GF89 Q16QG8 T1J6K7 A0A1L8DUT8 A0A1Y1M833 Q1HR80 A0A2G8K3X9 E2AVC2 V4C6M3 A7RY76 A0A2P6KSF9 A0A3M7RN27 A0A0P4VYP9 A0A2P8XW72 A0A0V0G774 A0A0P5QJ23 A0A0N8C2N8 A0A0P5QGX7 A0A0P5HM13 A0A0P5RA20 A0A0P5MNN2 A0A0P5S2M6 A0A0P5FQ20 A0A0N8BPN4 A0A0P6HT18 A0A0P5JJK9 A0A0P5NJU5 A0A0P6D3Q1 A0A0P5RVH1 A0A0P5KVK1 A0A0P6EC03 A0A195EEQ8 A0A310SFU5 E9GLS7 A0A0P5LFK4 F4WQY8 K1Q1U5 E0VCW2 A0A0P5GA51 A0A158NTL7 A0A0P5J7G8 A0A0P6E4D8 A0A069DPR8 A0A1D2NJ59 A0A0P5KJ71 A0A0P5IM56 A0A0P5Z6D9 C3XVV5 A0A0P5BUM2 A0A026VZ16 A0A0P5KW66 A0A023GNY9 A0A0P5KZR6 G3MR58 A0A0P5AHE6 T1KBS0 A0A0P5YG89 A0A0P5E293 A0A0P5YUI7 A0A224XV21 A0A2B4S7Y4 A0A0P5X1W3 A0A0P5W592 A0A0P5Z889 A0A0P5B2Q3 A0A0N8D179 A0A0P5VW13 A0A3R7LZU9 A0A0P5SX60 A0A0N7ZR73 A0A0P5EC66 A0A0P4YNZ5 A0A0P5SW50 A0A0P6D029 A0A1B0EUN5 A0A0P4Z434 A0A0P5AC32 A0A0N8D8J1 A0A0P6BWX9 A0A2H1BVZ3
Ontologies
PATHWAY
Topology
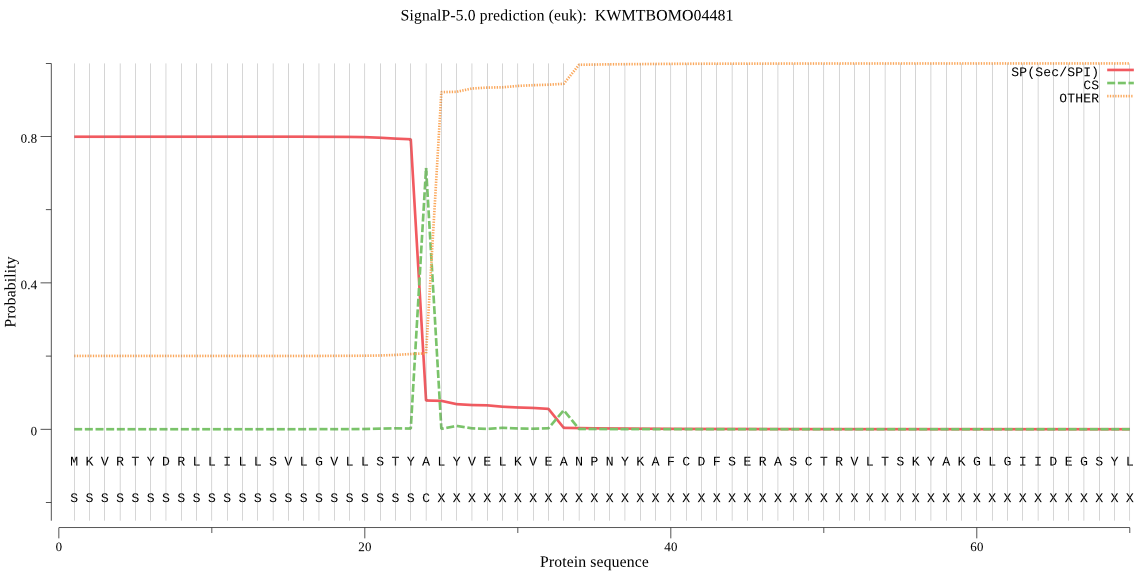
SignalP
Position: 1 - 24,
Likelihood: 0.799441
Length:
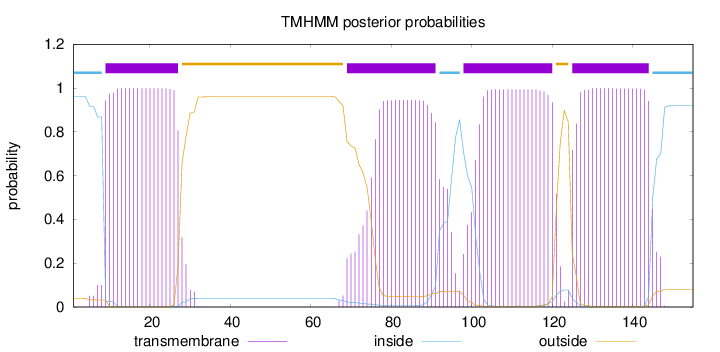
155
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
80.56606
Exp number, first 60 AAs:
19.63021
Total prob of N-in:
0.96175
POSSIBLE N-term signal
sequence
inside
1 - 8
TMhelix
9 - 27
outside
28 - 68
TMhelix
69 - 91
inside
92 - 97
TMhelix
98 - 120
outside
121 - 124
TMhelix
125 - 144
inside
145 - 155
Population Genetic Test Statistics
Pi
42.801864
Theta
158.681022
Tajima's D
-1.102553
CLR
10.433385
CSRT
0.117194140292985
Interpretation
Uncertain
