Gene
KWMTBOMO04478
Pre Gene Modal
BGIBMGA005367
Annotation
PREDICTED:_cell_cycle_checkpoint_protein_RAD17_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.094
Sequence
CDS
ATGCTAAAAAAGCATAAGACCTGGTTTAAACCAATGTTTGATTTTGATGAAACTGAGCCTCCACCGCAGAAGCTTAAAAGTGATGTAAACAAACAAGGTACGAGTTCACCAGCCAAGTTTTCAAAATCGAACACTATTAAACCAATGATTTTTAATATCATTGATCAAAACCACATTCGGAATTTTGATCCGGTCAATATCAATGACTTGGCCATACACAATAAAAAGGTTCAAGAAGTTGAAGATTGGATATTAGGTAACATAAAGAAGAATACAAGTGAAATGTTGCTCCTTACTGGACCAGTCGGCTGCGGCAAGAGTGTTACAGTGCTGATTATAGCTAAAAAACATAATGTAAAAATTACAGAGTGGATTACACCTATTGATATTGAATTCCCTAATGATAATGGAGAGTTTGAATTTAAAGTTAAACAATCTACTCAATTTTTGGATTTTATTTTAAATGCAGCCAACTTTACTTCACTGCTTGATAATAATGACAGAAAACTGGTGTTAGTAGAAGATTTCCCAAACACCTTTATGAGAACCCCTTCAGAATTAGTTTATGTATTACAACAATACAAACAGAGGGCAAAATCGCCAATTGTTTTCATTTGTTCTGAATCAAGTACAGACAAAAAGACCACCAGTGGAAACTTATTTAGTGATCAACTGAAGAGCCAACTGAATATTCATCATATTAGTTTTAATGGTGTTTCGACAACCGGCTTACGGTCTGCATTAAAACGAATGGCAGAGATAATAAGTAAAAAGTACAAATCAATGTACAATGCTCCTACAAGTGACCTGATTGAATGTGTTGTCAACTCTTCAGCAGGCGACGTCAGGTCAGCCGTAACAAATTTGCATTTTGCATGTTTAAAAGGTACTGATCAAAGTTTAGAAACCAGTATAATAACGGAAAAATACCAGAAAACTAAAACTAAGAAGAAAAAACCAAGTAGCAAATTTACTTCAATAGGAAAAGATCAAAGTATCAATATTCTTCATGGAGTTGGAAGAGTTTTAAATCCTAGAGTTGTTGATGTTAACGGAAGAAGTCAATTTACACACTCTCCAAGAGAAATAGTAGAACAATTTATTTCTCAACCAAGCTCTTTTGTGAATTTTATTGAAGAAAACTACATGCCACATTTCTCAAATTCAGCTCAAATAGATAGAGCTGCTTCTACTCTTAGCGACGCTGTCTTCATGCTTGCGGAATGGAGGGAAAAAGTGTGTCAAGAATATGGGTTATATTGCGCAGTAGCTGGTCTAATGCTGGCTAACAAAGACCCTGTCTCAGCTTGGAATCCTGTACGTGGGCCTAAAAATATGAAAATCACTTACCCGGTCCCTTCAGAATTGCAACTTTTAGAAGAAAATTATTTGTACAAAGGAAAAACTCTTGTTTCTGATTACCAAACCTTTTGTAAAATTATTAACAGTAATTATGATTAA
Protein
MLKKHKTWFKPMFDFDETEPPPQKLKSDVNKQGTSSPAKFSKSNTIKPMIFNIIDQNHIRNFDPVNINDLAIHNKKVQEVEDWILGNIKKNTSEMLLLTGPVGCGKSVTVLIIAKKHNVKITEWITPIDIEFPNDNGEFEFKVKQSTQFLDFILNAANFTSLLDNNDRKLVLVEDFPNTFMRTPSELVYVLQQYKQRAKSPIVFICSESSTDKKTTSGNLFSDQLKSQLNIHHISFNGVSTTGLRSALKRMAEIISKKYKSMYNAPTSDLIECVVNSSAGDVRSAVTNLHFACLKGTDQSLETSIITEKYQKTKTKKKKPSSKFTSIGKDQSINILHGVGRVLNPRVVDVNGRSQFTHSPREIVEQFISQPSSFVNFIEENYMPHFSNSAQIDRAASTLSDAVFMLAEWREKVCQEYGLYCAVAGLMLANKDPVSAWNPVRGPKNMKITYPVPSELQLLEENYLYKGKTLVSDYQTFCKIINSNYD
Summary
Uniprot
A0A2H1VV39
A0A2A4JZI0
A0A3S2M0E2
A0A194QSF4
A0A2A4K0G2
A0A212ERE7
+ More
A0A194PNX6 B4JH55 B4LXB1 A0A1A9XC36 A0A1B0BHR7 A0A1A9VIJ7 A0A1B0FLJ0 B4KA38 A0A1A9WCB6 A0A1I8MN85 A0A1B0A9Z7 B4NJU3 A0A1I8P3C9 B4GNP9 B5DXR6 A0A0L0CC04 B4QZS6 B3P3M5 B3LW20 A0A0M5IZG9 A0A3B0JDB0 B4HEK1 Q7KND1 B4PSQ5 B8A3U8 O96532 A0A1W4V5S7 Q8SYV6 A0A0K8VAE2 A0A0A1XQZ8 A0A1Q3F982 A0A1B0GJ33 A0A034VVM9 B0W9K3 W8BY80 A0A034VXA3 A0A182HBR8 A0A0N8ES88 Q171F4 A0A1W4X1R9 A0A1Y1LRS7 A0A087ZR47 A0A1Y9G9I6 W5JMK7 A0A2A3E7J2 A0A310SVF5 A0A0M9A9I7 A0A1Y9IVK9 A0A182PJG1 A0A182MHR9 A0A0L7QLD9 A0A154PHH6 D6WXK2 A0A0J7KN03 A0A182KAT6 E2AHQ6 A0A182IBG4 A0A1S4GYX9 A0A182U0S0 A0A1B6CHY6 A0A232FC99 K7JF94 Q7Q8E2 A0A158NY25 A0A182V0Q6 A0A1J1HKG5 A0A1B6H1J3 A0A1B6JWH1 A0A182RK17 A0A1B6J882 F4WLQ3 A0A182QA32 A0A1B6LNP8 A0A182NGT0 E2BA60 A0A151II21 A0A1J1HFZ3 A0A182J0E3 A0A3Q3F1R3 A0A151JTH2 A0A1S3K9T0 A0A1S3KA59 A0A151X0W5 N6T698 A0A1S3JCP1 J9KAX6 A0A2S2NMK9 A0A0C9R170
A0A194PNX6 B4JH55 B4LXB1 A0A1A9XC36 A0A1B0BHR7 A0A1A9VIJ7 A0A1B0FLJ0 B4KA38 A0A1A9WCB6 A0A1I8MN85 A0A1B0A9Z7 B4NJU3 A0A1I8P3C9 B4GNP9 B5DXR6 A0A0L0CC04 B4QZS6 B3P3M5 B3LW20 A0A0M5IZG9 A0A3B0JDB0 B4HEK1 Q7KND1 B4PSQ5 B8A3U8 O96532 A0A1W4V5S7 Q8SYV6 A0A0K8VAE2 A0A0A1XQZ8 A0A1Q3F982 A0A1B0GJ33 A0A034VVM9 B0W9K3 W8BY80 A0A034VXA3 A0A182HBR8 A0A0N8ES88 Q171F4 A0A1W4X1R9 A0A1Y1LRS7 A0A087ZR47 A0A1Y9G9I6 W5JMK7 A0A2A3E7J2 A0A310SVF5 A0A0M9A9I7 A0A1Y9IVK9 A0A182PJG1 A0A182MHR9 A0A0L7QLD9 A0A154PHH6 D6WXK2 A0A0J7KN03 A0A182KAT6 E2AHQ6 A0A182IBG4 A0A1S4GYX9 A0A182U0S0 A0A1B6CHY6 A0A232FC99 K7JF94 Q7Q8E2 A0A158NY25 A0A182V0Q6 A0A1J1HKG5 A0A1B6H1J3 A0A1B6JWH1 A0A182RK17 A0A1B6J882 F4WLQ3 A0A182QA32 A0A1B6LNP8 A0A182NGT0 E2BA60 A0A151II21 A0A1J1HFZ3 A0A182J0E3 A0A3Q3F1R3 A0A151JTH2 A0A1S3K9T0 A0A1S3KA59 A0A151X0W5 N6T698 A0A1S3JCP1 J9KAX6 A0A2S2NMK9 A0A0C9R170
Pubmed
26354079
22118469
17994087
25315136
15632085
26108605
+ More
9878245 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25830018 25348373 24495485 26483478 26999592 17510324 28004739 20920257 23761445 18362917 19820115 20798317 12364791 28648823 20075255 21347285 21719571 23537049
9878245 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25830018 25348373 24495485 26483478 26999592 17510324 28004739 20920257 23761445 18362917 19820115 20798317 12364791 28648823 20075255 21347285 21719571 23537049
EMBL
ODYU01004455
SOQ44342.1
NWSH01000327
PCG77407.1
RSAL01000088
RVE48211.1
+ More
KQ461154 KPJ08417.1 PCG77408.1 AGBW02013068 OWR44024.1 KQ459599 KPI94449.1 CH916369 EDV92746.1 CH940650 EDW67789.1 JXJN01014542 CCAG010021492 CH933806 EDW16713.2 CH964272 EDW85055.1 CH479186 EDW39375.1 CM000070 EDY67507.3 JRES01000720 KNC28974.1 CM000364 EDX12924.1 CH954181 EDV48913.1 CH902617 EDV41553.1 CP012526 ALC45518.1 OUUW01000005 SPP80364.1 CH480815 EDW42158.1 AF076839 AF124503 AAC95521.1 AAD31692.1 CM000160 EDW97551.1 BT056240 ACL68687.1 AE014297 AAF55086.2 AY071291 AAL48913.1 GDHF01016527 JAI35787.1 GBXI01000920 JAD13372.1 GFDL01010904 JAV24141.1 AJWK01019254 GAKP01012453 JAC46499.1 DS231864 EDS40228.1 GAMC01002248 JAC04308.1 GAKP01012452 JAC46500.1 JXUM01126568 KQ567082 KXJ69627.1 GDUN01000146 JAN95773.1 CH477456 EAT40637.1 GEZM01048838 JAV76359.1 ADMH02000616 ETN65612.1 KZ288344 PBC27717.1 KQ759804 OAD62799.1 KQ435709 KOX79987.1 AXCM01008036 KQ414924 KOC59428.1 KQ434907 KZC11301.1 KQ971361 EFA07969.1 LBMM01005167 KMQ91768.1 GL439579 EFN67036.1 APCN01000686 AAAB01008944 GEDC01024288 JAS13010.1 NNAY01000459 OXU28242.1 AAZX01008339 EAA10117.2 ADTU01002955 CVRI01000002 CRK86950.1 GECZ01005752 GECZ01001228 JAS64017.1 JAS68541.1 GECU01004170 JAT03537.1 GECU01012336 JAS95370.1 GL888212 EGI64881.1 AXCN02002201 GEBQ01014758 JAT25219.1 GL446678 EFN87402.1 KQ977542 KYN02061.1 CRK86949.1 KQ981972 KYN31643.1 KQ982600 KYQ54026.1 APGK01050708 KB741176 ENN73193.1 ABLF02024419 GGMR01005749 MBY18368.1 GBYB01006642 JAG76409.1
KQ461154 KPJ08417.1 PCG77408.1 AGBW02013068 OWR44024.1 KQ459599 KPI94449.1 CH916369 EDV92746.1 CH940650 EDW67789.1 JXJN01014542 CCAG010021492 CH933806 EDW16713.2 CH964272 EDW85055.1 CH479186 EDW39375.1 CM000070 EDY67507.3 JRES01000720 KNC28974.1 CM000364 EDX12924.1 CH954181 EDV48913.1 CH902617 EDV41553.1 CP012526 ALC45518.1 OUUW01000005 SPP80364.1 CH480815 EDW42158.1 AF076839 AF124503 AAC95521.1 AAD31692.1 CM000160 EDW97551.1 BT056240 ACL68687.1 AE014297 AAF55086.2 AY071291 AAL48913.1 GDHF01016527 JAI35787.1 GBXI01000920 JAD13372.1 GFDL01010904 JAV24141.1 AJWK01019254 GAKP01012453 JAC46499.1 DS231864 EDS40228.1 GAMC01002248 JAC04308.1 GAKP01012452 JAC46500.1 JXUM01126568 KQ567082 KXJ69627.1 GDUN01000146 JAN95773.1 CH477456 EAT40637.1 GEZM01048838 JAV76359.1 ADMH02000616 ETN65612.1 KZ288344 PBC27717.1 KQ759804 OAD62799.1 KQ435709 KOX79987.1 AXCM01008036 KQ414924 KOC59428.1 KQ434907 KZC11301.1 KQ971361 EFA07969.1 LBMM01005167 KMQ91768.1 GL439579 EFN67036.1 APCN01000686 AAAB01008944 GEDC01024288 JAS13010.1 NNAY01000459 OXU28242.1 AAZX01008339 EAA10117.2 ADTU01002955 CVRI01000002 CRK86950.1 GECZ01005752 GECZ01001228 JAS64017.1 JAS68541.1 GECU01004170 JAT03537.1 GECU01012336 JAS95370.1 GL888212 EGI64881.1 AXCN02002201 GEBQ01014758 JAT25219.1 GL446678 EFN87402.1 KQ977542 KYN02061.1 CRK86949.1 KQ981972 KYN31643.1 KQ982600 KYQ54026.1 APGK01050708 KB741176 ENN73193.1 ABLF02024419 GGMR01005749 MBY18368.1 GBYB01006642 JAG76409.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000007151
UP000053268
UP000001070
+ More
UP000008792 UP000092443 UP000092460 UP000078200 UP000092444 UP000009192 UP000091820 UP000095301 UP000092445 UP000007798 UP000095300 UP000008744 UP000001819 UP000037069 UP000000304 UP000008711 UP000007801 UP000092553 UP000268350 UP000001292 UP000002282 UP000000803 UP000192221 UP000092461 UP000002320 UP000069940 UP000249989 UP000008820 UP000192223 UP000005203 UP000069272 UP000000673 UP000242457 UP000053105 UP000075920 UP000075885 UP000075883 UP000053825 UP000076502 UP000007266 UP000036403 UP000075881 UP000000311 UP000075840 UP000075902 UP000215335 UP000002358 UP000007062 UP000005205 UP000075903 UP000183832 UP000075900 UP000007755 UP000075886 UP000075884 UP000008237 UP000078542 UP000075880 UP000261660 UP000078541 UP000085678 UP000075809 UP000019118 UP000007819
UP000008792 UP000092443 UP000092460 UP000078200 UP000092444 UP000009192 UP000091820 UP000095301 UP000092445 UP000007798 UP000095300 UP000008744 UP000001819 UP000037069 UP000000304 UP000008711 UP000007801 UP000092553 UP000268350 UP000001292 UP000002282 UP000000803 UP000192221 UP000092461 UP000002320 UP000069940 UP000249989 UP000008820 UP000192223 UP000005203 UP000069272 UP000000673 UP000242457 UP000053105 UP000075920 UP000075885 UP000075883 UP000053825 UP000076502 UP000007266 UP000036403 UP000075881 UP000000311 UP000075840 UP000075902 UP000215335 UP000002358 UP000007062 UP000005205 UP000075903 UP000183832 UP000075900 UP000007755 UP000075886 UP000075884 UP000008237 UP000078542 UP000075880 UP000261660 UP000078541 UP000085678 UP000075809 UP000019118 UP000007819
Interpro
ProteinModelPortal
A0A2H1VV39
A0A2A4JZI0
A0A3S2M0E2
A0A194QSF4
A0A2A4K0G2
A0A212ERE7
+ More
A0A194PNX6 B4JH55 B4LXB1 A0A1A9XC36 A0A1B0BHR7 A0A1A9VIJ7 A0A1B0FLJ0 B4KA38 A0A1A9WCB6 A0A1I8MN85 A0A1B0A9Z7 B4NJU3 A0A1I8P3C9 B4GNP9 B5DXR6 A0A0L0CC04 B4QZS6 B3P3M5 B3LW20 A0A0M5IZG9 A0A3B0JDB0 B4HEK1 Q7KND1 B4PSQ5 B8A3U8 O96532 A0A1W4V5S7 Q8SYV6 A0A0K8VAE2 A0A0A1XQZ8 A0A1Q3F982 A0A1B0GJ33 A0A034VVM9 B0W9K3 W8BY80 A0A034VXA3 A0A182HBR8 A0A0N8ES88 Q171F4 A0A1W4X1R9 A0A1Y1LRS7 A0A087ZR47 A0A1Y9G9I6 W5JMK7 A0A2A3E7J2 A0A310SVF5 A0A0M9A9I7 A0A1Y9IVK9 A0A182PJG1 A0A182MHR9 A0A0L7QLD9 A0A154PHH6 D6WXK2 A0A0J7KN03 A0A182KAT6 E2AHQ6 A0A182IBG4 A0A1S4GYX9 A0A182U0S0 A0A1B6CHY6 A0A232FC99 K7JF94 Q7Q8E2 A0A158NY25 A0A182V0Q6 A0A1J1HKG5 A0A1B6H1J3 A0A1B6JWH1 A0A182RK17 A0A1B6J882 F4WLQ3 A0A182QA32 A0A1B6LNP8 A0A182NGT0 E2BA60 A0A151II21 A0A1J1HFZ3 A0A182J0E3 A0A3Q3F1R3 A0A151JTH2 A0A1S3K9T0 A0A1S3KA59 A0A151X0W5 N6T698 A0A1S3JCP1 J9KAX6 A0A2S2NMK9 A0A0C9R170
A0A194PNX6 B4JH55 B4LXB1 A0A1A9XC36 A0A1B0BHR7 A0A1A9VIJ7 A0A1B0FLJ0 B4KA38 A0A1A9WCB6 A0A1I8MN85 A0A1B0A9Z7 B4NJU3 A0A1I8P3C9 B4GNP9 B5DXR6 A0A0L0CC04 B4QZS6 B3P3M5 B3LW20 A0A0M5IZG9 A0A3B0JDB0 B4HEK1 Q7KND1 B4PSQ5 B8A3U8 O96532 A0A1W4V5S7 Q8SYV6 A0A0K8VAE2 A0A0A1XQZ8 A0A1Q3F982 A0A1B0GJ33 A0A034VVM9 B0W9K3 W8BY80 A0A034VXA3 A0A182HBR8 A0A0N8ES88 Q171F4 A0A1W4X1R9 A0A1Y1LRS7 A0A087ZR47 A0A1Y9G9I6 W5JMK7 A0A2A3E7J2 A0A310SVF5 A0A0M9A9I7 A0A1Y9IVK9 A0A182PJG1 A0A182MHR9 A0A0L7QLD9 A0A154PHH6 D6WXK2 A0A0J7KN03 A0A182KAT6 E2AHQ6 A0A182IBG4 A0A1S4GYX9 A0A182U0S0 A0A1B6CHY6 A0A232FC99 K7JF94 Q7Q8E2 A0A158NY25 A0A182V0Q6 A0A1J1HKG5 A0A1B6H1J3 A0A1B6JWH1 A0A182RK17 A0A1B6J882 F4WLQ3 A0A182QA32 A0A1B6LNP8 A0A182NGT0 E2BA60 A0A151II21 A0A1J1HFZ3 A0A182J0E3 A0A3Q3F1R3 A0A151JTH2 A0A1S3K9T0 A0A1S3KA59 A0A151X0W5 N6T698 A0A1S3JCP1 J9KAX6 A0A2S2NMK9 A0A0C9R170
Ontologies
GO
PANTHER
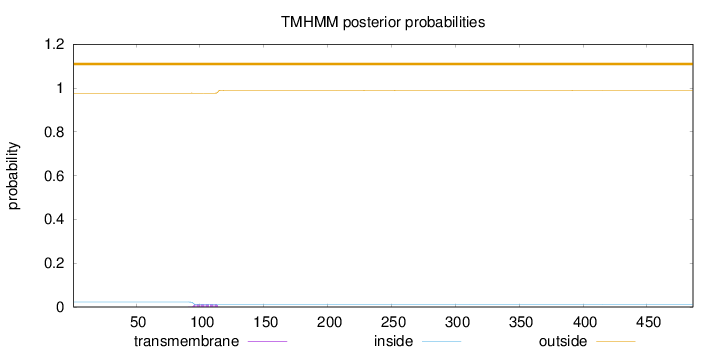
Topology
Length:
486
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.27334
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02335
outside
1 - 486
Population Genetic Test Statistics
Pi
19.775977
Theta
157.54135
Tajima's D
0.7086
CLR
0.571306
CSRT
0.578171091445428
Interpretation
Uncertain
