Gene
KWMTBOMO04474
Pre Gene Modal
BGIBMGA005370
Annotation
cell_cycle_checkpoint_kinase_2_[Bombyx_mori]
Full name
Ovarian-specific serine/threonine-protein kinase Lok
Alternative Name
Protein loki
dMNK
dMNK
Location in the cell
Nuclear Reliability : 2.832
Sequence
CDS
ATGGGGGACGATCCAATACACGATAGCGAAAACACTCAAACACAAACCCAAAACTCACAAGTAGATTGGAGTCAGCAAAATACACCAGCCACAATTCCAAATATTTGGGGAAGATTGTATTCTACAAAGGTATCAATTAATGGTAAATATTGTTGGAAACAAGAATATCAACCTGAATATATCGATCTTATACAACCCGAATTCAGCTTGGGAAGATCATTCAACTGCACTTTCATAATGAGAAAAGACAATATAAAAGAGAGTATTATAAGGAATGTGAGCAAGCTACACTTTGTTATTAAGAGAAATCTATCTGAACTTCTAAGCCCTGCTATAATAACAGACCTGTCACACAATGGTACCTTTGTGAATGGCGAAGACATAGGTAAAGGAAACAGTAGAGTTCTAGATGACAATGATGTAATATCAGTTACTCATCGTATTGTTAAAATTTTTATATTTAAAGACTTATTGAAAAACGAACAAGACCAAGTACCTAAAGAAATTTCACAAAAGTATTATATCTCAAGGGTTTTAGGGCAGGGGGCTTGCGGCCTTGTTAAATTAGTTTATGACAAGATAAAATGCACCAAGCATGCCATGAAAATTATAAAAAAAAGTAGACTTACAAATGGTCAAATTAATCACATAAATAAGCCCGCTAAAATTATGAACGAGATCAATATAATGAAAGCACTCAGACATCCATGCATTATTTCAATAGAAGAAGTATTTGACTCTCGCGATGCTGTGTATATTATTTTAGAACTCATGGAAGGCGGTGAACTGTTTGATAGGATAACAAAATTTAGTCACTTGACAGAACCATTAACACGGTTTATTTTCCGACAAATGGTACTTGCAGTCAAGTACTTGCACTCTCAGGGTATAACTCATCGGGATTTGAAGCCTGAGAATGTCCTGTTGGAAGGTGACGAAGACGAAACATTAGTGAAAATTACTGACTTTGGACTGAGTAAGTTTGTAGGAGAGGACAGCTTTATGAAGACAATGTGTGGGACACCATTGTATCTGGCTCCAGAAGTGCTCAGAGCTAATGGACAAAATACTTATGGACCAGAGGTTGATGTTTGGAGTCTAGGAGTCATATTCTTTGTATGCCTTGTGGGTTATTTACCCTTCTCGCACGACTACAAAGAGATGTCACTCAAGAACCAGATATTAAATGGACAATACAAGTTCTCACAGGCTCACTGGAAGAATATAAGCTTGCAGGCCAAGCTGCTTATGAAAAGAATGCTCACAGTGCAAGTCAGCAGGAGGATAACCCTTGATCAGATACTGAATCATGCTTGGATGCAGGACGTTGATACGATCATAAGGGTTGAGAAACTTCTATCACACATCCGCTCTAAGGGCATGAAACAGGCGACAAATAGCACCACCGGTGACGGCGACGACAATGTCCGTAACAGCACGGTGGCCATAATACAAGTGGTAGCCGCGGGTAAGCGAGCCCTCAGCGACACGTTCTGCGCCACCGAACCGAACGCAAAAAAAATTAAAATAGCCTTGACCAATGACGACGAAACTAGTTCCACAAACAGTTATTGCTCAAATGCTTAA
Protein
MGDDPIHDSENTQTQTQNSQVDWSQQNTPATIPNIWGRLYSTKVSINGKYCWKQEYQPEYIDLIQPEFSLGRSFNCTFIMRKDNIKESIIRNVSKLHFVIKRNLSELLSPAIITDLSHNGTFVNGEDIGKGNSRVLDDNDVISVTHRIVKIFIFKDLLKNEQDQVPKEISQKYYISRVLGQGACGLVKLVYDKIKCTKHAMKIIKKSRLTNGQINHINKPAKIMNEINIMKALRHPCIISIEEVFDSRDAVYIILELMEGGELFDRITKFSHLTEPLTRFIFRQMVLAVKYLHSQGITHRDLKPENVLLEGDEDETLVKITDFGLSKFVGEDSFMKTMCGTPLYLAPEVLRANGQNTYGPEVDVWSLGVIFFVCLVGYLPFSHDYKEMSLKNQILNGQYKFSQAHWKNISLQAKLLMKRMLTVQVSRRITLDQILNHAWMQDVDTIIRVEKLLSHIRSKGMKQATNSTTGDGDDNVRNSTVAIIQVVAAGKRALSDTFCATEPNAKKIKIALTNDDETSSTNSYCSNA
Summary
Description
May have a role in germline establishment.
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Similarity
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily. CAMK Ser/Thr protein kinase family. CDS1 subfamily.
Belongs to the ubiquitin-conjugating enzyme family.
Belongs to the protein kinase superfamily. CAMK Ser/Thr protein kinase family. CDS1 subfamily.
Belongs to the ubiquitin-conjugating enzyme family.
Keywords
Alternative splicing
ATP-binding
Complete proteome
Kinase
Nucleotide-binding
Nucleus
Reference proteome
Transferase
Feature
chain Ovarian-specific serine/threonine-protein kinase Lok
splice variant In isoform Short.
splice variant In isoform Short.
Uniprot
H9J778
Q5R1T9
A0A2A4K0W4
A0A2H1X294
A0A212FBW0
A0A3S2M0C9
+ More
A0A194QTU5 A0A194PLY2 A0A2A4K186 A0A1I8MT58 A0A034WRB7 A0A0L0C9G7 A0A0K8UI60 A0A1I8P361 A0A1I8P303 A0A0A1X8Z4 W8BDL4 A0A067QEX6 A0A0Q5VK69 B3NL10 O61267 O61267-2 A4V0X0 A0A0R1DPJ5 A0A1W4VYG8 A0A0J9R4V2 A0A0Q9XBV4 B4JQ87 A0A1W4VK90 B4PBH1 A0A0Q9X4P6 B4N7N9 B4KEP1 B3ML32 A0A0P8ZFI1 B4M9L4 V4C6S6 A0A1S3HKX9 A0A1A9W2P4 B4I626 A0A2T7PDM7 A0A0M9AAQ0 A0A2J7RJL5 A0A3B0JV41 A0A0M3QU81 A0A0B6XZ20 Q29KK4 B4GT76 A0A0R3NUJ5 A0A087ZR28 Q171S5 A0A2Z5U631 K1R1E4 A0A2A3E930 A0A1Q3F563 A0A210R6F6 A0A1Q3F527 A0A1Q3F521 A0A154PFN3 A0A1Q3F582 B0WQ25 A0A1B0GDY8 A0A1A9VM20 A0A2C9JI89 A0A232EYX4 K7ISU1 A0A0L8G8I3 A0A1A9Y7G5 A0A1A9UNG1 A0A1B0BTC4 A0A1B0BKJ7 T1IRU8 A0A1B0A5M9 A0A1Q3F5F1 A0A2G8K946 A0A1B0FRF0 A0A3L8SBJ6 A7RVB0 A0A0J7NZB3 G7Y516 A0A0R3VXU3 H0ZC01 W5JFI3 A0A091N9K7 A0A1B0AHF7 A0A093QGW9 A0A3M7TA35 A0A093HKE0 A0A091ENI5 Q98TW0 A0A091HHU0 A0A2M4CYM9 Q9I8V3 A0A093PI42 A0A182WGM0 A0A182MQ34 A0A091GBF5 A0A2M4CYG6 A0A182HRU3
A0A194QTU5 A0A194PLY2 A0A2A4K186 A0A1I8MT58 A0A034WRB7 A0A0L0C9G7 A0A0K8UI60 A0A1I8P361 A0A1I8P303 A0A0A1X8Z4 W8BDL4 A0A067QEX6 A0A0Q5VK69 B3NL10 O61267 O61267-2 A4V0X0 A0A0R1DPJ5 A0A1W4VYG8 A0A0J9R4V2 A0A0Q9XBV4 B4JQ87 A0A1W4VK90 B4PBH1 A0A0Q9X4P6 B4N7N9 B4KEP1 B3ML32 A0A0P8ZFI1 B4M9L4 V4C6S6 A0A1S3HKX9 A0A1A9W2P4 B4I626 A0A2T7PDM7 A0A0M9AAQ0 A0A2J7RJL5 A0A3B0JV41 A0A0M3QU81 A0A0B6XZ20 Q29KK4 B4GT76 A0A0R3NUJ5 A0A087ZR28 Q171S5 A0A2Z5U631 K1R1E4 A0A2A3E930 A0A1Q3F563 A0A210R6F6 A0A1Q3F527 A0A1Q3F521 A0A154PFN3 A0A1Q3F582 B0WQ25 A0A1B0GDY8 A0A1A9VM20 A0A2C9JI89 A0A232EYX4 K7ISU1 A0A0L8G8I3 A0A1A9Y7G5 A0A1A9UNG1 A0A1B0BTC4 A0A1B0BKJ7 T1IRU8 A0A1B0A5M9 A0A1Q3F5F1 A0A2G8K946 A0A1B0FRF0 A0A3L8SBJ6 A7RVB0 A0A0J7NZB3 G7Y516 A0A0R3VXU3 H0ZC01 W5JFI3 A0A091N9K7 A0A1B0AHF7 A0A093QGW9 A0A3M7TA35 A0A093HKE0 A0A091ENI5 Q98TW0 A0A091HHU0 A0A2M4CYM9 Q9I8V3 A0A093PI42 A0A182WGM0 A0A182MQ34 A0A091GBF5 A0A2M4CYG6 A0A182HRU3
EC Number
2.7.11.1
Pubmed
19121390
22118469
26354079
25315136
25348373
26108605
+ More
25830018 24495485 24845553 17994087 9507063 10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867 17550304 22936249 23254933 26383154 15632085 17510324 26760975 22992520 28812685 15562597 28648823 20075255 29023486 30282656 17615350 22023798 20360741 20920257 23761445 30375419 10793133
25830018 24495485 24845553 17994087 9507063 10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867 17550304 22936249 23254933 26383154 15632085 17510324 26760975 22992520 28812685 15562597 28648823 20075255 29023486 30282656 17615350 22023798 20360741 20920257 23761445 30375419 10793133
EMBL
BABH01013683
BABH01013684
AB194683
BAD74191.1
NWSH01000327
PCG77403.1
+ More
ODYU01012852 SOQ59358.1 AGBW02009268 OWR51215.1 RSAL01000088 RVE48215.1 KQ461154 KPJ08420.1 KQ459599 KPI94446.1 PCG77402.1 GAKP01000821 JAC58131.1 JRES01000736 KNC28882.1 GDHF01031755 GDHF01025940 GDHF01002665 GDHF01002632 JAI20559.1 JAI26374.1 JAI49649.1 JAI49682.1 GBXI01006946 GBXI01006052 JAD07346.1 JAD08240.1 GAMC01007160 JAB99395.1 KK853689 KDQ97283.1 CH954179 KQS61847.1 EDV54588.1 AB007821 AB007822 U87984 AE014134 AY070549 AAN11063.1 CM000158 KRJ99215.1 CM002910 KMY91118.1 CH933807 KRG02970.1 CH916372 EDV99067.1 EDW90485.1 CH964214 KRF99271.1 EDW80378.1 EDW11920.1 CH902620 EDV31650.1 KPU73526.1 CH940654 EDW57890.1 KB201305 ESO97334.1 CH480822 EDW55832.1 PZQS01000004 PVD31511.1 KQ435709 KOX79956.1 NEVH01002992 PNF41023.1 OUUW01000010 SPP85965.1 CP012523 ALC40203.1 HACG01002243 CEK49108.1 CH379061 EAL33170.2 CH479189 EDW25585.1 KRT04792.1 CH477447 EAT40742.1 FX985821 BBA93708.1 JH818716 EKC37339.1 KZ288344 PBC27699.1 GFDL01012376 JAV22669.1 NEDP02000123 OWF56643.1 GFDL01012378 JAV22667.1 GFDL01012383 JAV22662.1 KQ434893 KZC10673.1 GFDL01012382 JAV22663.1 DS232032 EDS32669.1 CCAG010012075 NNAY01001525 OXU23686.1 AAZX01006436 KQ423210 KOF73346.1 JXJN01020073 JXJN01015972 JH431388 GFDL01012250 JAV22795.1 MRZV01000774 PIK44483.1 CCAG010004927 QUSF01000030 RLV99732.1 DS469543 EDO44613.1 LBMM01000692 KMQ97735.1 DF142866 GAA48052.1 UYRS01001261 VDK24633.1 ABQF01029141 ADMH02001647 ETN61569.1 KK843751 KFP85712.1 KL671659 KFW83347.1 REGN01000043 RNA44946.1 KL206193 KFV79870.1 KK718897 KFO59498.1 AF326574 BC166130 AAG59884.1 AAI66130.1 KL217497 KFO95853.1 GGFL01006197 MBW70375.1 AF174295 AAF75829.1 KL225417 KFW71867.1 AXCM01002017 KL447264 KFO71512.1 GGFL01006196 MBW70374.1 APCN01001747
ODYU01012852 SOQ59358.1 AGBW02009268 OWR51215.1 RSAL01000088 RVE48215.1 KQ461154 KPJ08420.1 KQ459599 KPI94446.1 PCG77402.1 GAKP01000821 JAC58131.1 JRES01000736 KNC28882.1 GDHF01031755 GDHF01025940 GDHF01002665 GDHF01002632 JAI20559.1 JAI26374.1 JAI49649.1 JAI49682.1 GBXI01006946 GBXI01006052 JAD07346.1 JAD08240.1 GAMC01007160 JAB99395.1 KK853689 KDQ97283.1 CH954179 KQS61847.1 EDV54588.1 AB007821 AB007822 U87984 AE014134 AY070549 AAN11063.1 CM000158 KRJ99215.1 CM002910 KMY91118.1 CH933807 KRG02970.1 CH916372 EDV99067.1 EDW90485.1 CH964214 KRF99271.1 EDW80378.1 EDW11920.1 CH902620 EDV31650.1 KPU73526.1 CH940654 EDW57890.1 KB201305 ESO97334.1 CH480822 EDW55832.1 PZQS01000004 PVD31511.1 KQ435709 KOX79956.1 NEVH01002992 PNF41023.1 OUUW01000010 SPP85965.1 CP012523 ALC40203.1 HACG01002243 CEK49108.1 CH379061 EAL33170.2 CH479189 EDW25585.1 KRT04792.1 CH477447 EAT40742.1 FX985821 BBA93708.1 JH818716 EKC37339.1 KZ288344 PBC27699.1 GFDL01012376 JAV22669.1 NEDP02000123 OWF56643.1 GFDL01012378 JAV22667.1 GFDL01012383 JAV22662.1 KQ434893 KZC10673.1 GFDL01012382 JAV22663.1 DS232032 EDS32669.1 CCAG010012075 NNAY01001525 OXU23686.1 AAZX01006436 KQ423210 KOF73346.1 JXJN01020073 JXJN01015972 JH431388 GFDL01012250 JAV22795.1 MRZV01000774 PIK44483.1 CCAG010004927 QUSF01000030 RLV99732.1 DS469543 EDO44613.1 LBMM01000692 KMQ97735.1 DF142866 GAA48052.1 UYRS01001261 VDK24633.1 ABQF01029141 ADMH02001647 ETN61569.1 KK843751 KFP85712.1 KL671659 KFW83347.1 REGN01000043 RNA44946.1 KL206193 KFV79870.1 KK718897 KFO59498.1 AF326574 BC166130 AAG59884.1 AAI66130.1 KL217497 KFO95853.1 GGFL01006197 MBW70375.1 AF174295 AAF75829.1 KL225417 KFW71867.1 AXCM01002017 KL447264 KFO71512.1 GGFL01006196 MBW70374.1 APCN01001747
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053240
UP000053268
+ More
UP000095301 UP000037069 UP000095300 UP000027135 UP000008711 UP000000803 UP000002282 UP000192221 UP000009192 UP000001070 UP000007798 UP000007801 UP000008792 UP000030746 UP000085678 UP000091820 UP000001292 UP000245119 UP000053105 UP000235965 UP000268350 UP000092553 UP000001819 UP000008744 UP000005203 UP000008820 UP000005408 UP000242457 UP000242188 UP000076502 UP000002320 UP000092444 UP000078200 UP000076420 UP000215335 UP000002358 UP000053454 UP000092443 UP000092460 UP000092445 UP000230750 UP000276834 UP000001593 UP000036403 UP000046400 UP000282613 UP000007754 UP000000673 UP000053258 UP000276133 UP000053584 UP000052976 UP000054308 UP000054081 UP000075920 UP000075883 UP000053760 UP000075840
UP000095301 UP000037069 UP000095300 UP000027135 UP000008711 UP000000803 UP000002282 UP000192221 UP000009192 UP000001070 UP000007798 UP000007801 UP000008792 UP000030746 UP000085678 UP000091820 UP000001292 UP000245119 UP000053105 UP000235965 UP000268350 UP000092553 UP000001819 UP000008744 UP000005203 UP000008820 UP000005408 UP000242457 UP000242188 UP000076502 UP000002320 UP000092444 UP000078200 UP000076420 UP000215335 UP000002358 UP000053454 UP000092443 UP000092460 UP000092445 UP000230750 UP000276834 UP000001593 UP000036403 UP000046400 UP000282613 UP000007754 UP000000673 UP000053258 UP000276133 UP000053584 UP000052976 UP000054308 UP000054081 UP000075920 UP000075883 UP000053760 UP000075840
Interpro
Gene 3D
ProteinModelPortal
H9J778
Q5R1T9
A0A2A4K0W4
A0A2H1X294
A0A212FBW0
A0A3S2M0C9
+ More
A0A194QTU5 A0A194PLY2 A0A2A4K186 A0A1I8MT58 A0A034WRB7 A0A0L0C9G7 A0A0K8UI60 A0A1I8P361 A0A1I8P303 A0A0A1X8Z4 W8BDL4 A0A067QEX6 A0A0Q5VK69 B3NL10 O61267 O61267-2 A4V0X0 A0A0R1DPJ5 A0A1W4VYG8 A0A0J9R4V2 A0A0Q9XBV4 B4JQ87 A0A1W4VK90 B4PBH1 A0A0Q9X4P6 B4N7N9 B4KEP1 B3ML32 A0A0P8ZFI1 B4M9L4 V4C6S6 A0A1S3HKX9 A0A1A9W2P4 B4I626 A0A2T7PDM7 A0A0M9AAQ0 A0A2J7RJL5 A0A3B0JV41 A0A0M3QU81 A0A0B6XZ20 Q29KK4 B4GT76 A0A0R3NUJ5 A0A087ZR28 Q171S5 A0A2Z5U631 K1R1E4 A0A2A3E930 A0A1Q3F563 A0A210R6F6 A0A1Q3F527 A0A1Q3F521 A0A154PFN3 A0A1Q3F582 B0WQ25 A0A1B0GDY8 A0A1A9VM20 A0A2C9JI89 A0A232EYX4 K7ISU1 A0A0L8G8I3 A0A1A9Y7G5 A0A1A9UNG1 A0A1B0BTC4 A0A1B0BKJ7 T1IRU8 A0A1B0A5M9 A0A1Q3F5F1 A0A2G8K946 A0A1B0FRF0 A0A3L8SBJ6 A7RVB0 A0A0J7NZB3 G7Y516 A0A0R3VXU3 H0ZC01 W5JFI3 A0A091N9K7 A0A1B0AHF7 A0A093QGW9 A0A3M7TA35 A0A093HKE0 A0A091ENI5 Q98TW0 A0A091HHU0 A0A2M4CYM9 Q9I8V3 A0A093PI42 A0A182WGM0 A0A182MQ34 A0A091GBF5 A0A2M4CYG6 A0A182HRU3
A0A194QTU5 A0A194PLY2 A0A2A4K186 A0A1I8MT58 A0A034WRB7 A0A0L0C9G7 A0A0K8UI60 A0A1I8P361 A0A1I8P303 A0A0A1X8Z4 W8BDL4 A0A067QEX6 A0A0Q5VK69 B3NL10 O61267 O61267-2 A4V0X0 A0A0R1DPJ5 A0A1W4VYG8 A0A0J9R4V2 A0A0Q9XBV4 B4JQ87 A0A1W4VK90 B4PBH1 A0A0Q9X4P6 B4N7N9 B4KEP1 B3ML32 A0A0P8ZFI1 B4M9L4 V4C6S6 A0A1S3HKX9 A0A1A9W2P4 B4I626 A0A2T7PDM7 A0A0M9AAQ0 A0A2J7RJL5 A0A3B0JV41 A0A0M3QU81 A0A0B6XZ20 Q29KK4 B4GT76 A0A0R3NUJ5 A0A087ZR28 Q171S5 A0A2Z5U631 K1R1E4 A0A2A3E930 A0A1Q3F563 A0A210R6F6 A0A1Q3F527 A0A1Q3F521 A0A154PFN3 A0A1Q3F582 B0WQ25 A0A1B0GDY8 A0A1A9VM20 A0A2C9JI89 A0A232EYX4 K7ISU1 A0A0L8G8I3 A0A1A9Y7G5 A0A1A9UNG1 A0A1B0BTC4 A0A1B0BKJ7 T1IRU8 A0A1B0A5M9 A0A1Q3F5F1 A0A2G8K946 A0A1B0FRF0 A0A3L8SBJ6 A7RVB0 A0A0J7NZB3 G7Y516 A0A0R3VXU3 H0ZC01 W5JFI3 A0A091N9K7 A0A1B0AHF7 A0A093QGW9 A0A3M7TA35 A0A093HKE0 A0A091ENI5 Q98TW0 A0A091HHU0 A0A2M4CYM9 Q9I8V3 A0A093PI42 A0A182WGM0 A0A182MQ34 A0A091GBF5 A0A2M4CYG6 A0A182HRU3
PDB
3I6U
E-value=7.19117e-93,
Score=870
Ontologies
GO
GO:0004672
GO:0005524
GO:0006282
GO:0005634
GO:0050321
GO:0044773
GO:0006919
GO:0051091
GO:0072332
GO:0035234
GO:0071480
GO:0030717
GO:0071481
GO:0004674
GO:0008630
GO:0045944
GO:0006974
GO:0006915
GO:0000077
GO:0016607
GO:0007281
GO:0006468
GO:0016021
GO:0005516
GO:0005737
GO:0018105
GO:0035556
GO:0046777
GO:0004683
GO:0009931
GO:0050821
GO:0001302
GO:0006978
GO:0006975
GO:0042803
GO:0042771
GO:0006302
GO:0019901
GO:0016605
GO:0044257
GO:0090307
GO:0042176
GO:0072428
GO:0000086
GO:0045893
GO:0000781
GO:0005794
GO:0031625
GO:0005515
GO:0006310
GO:0003676
GO:0006418
GO:0009058
GO:0016742
GO:0003824
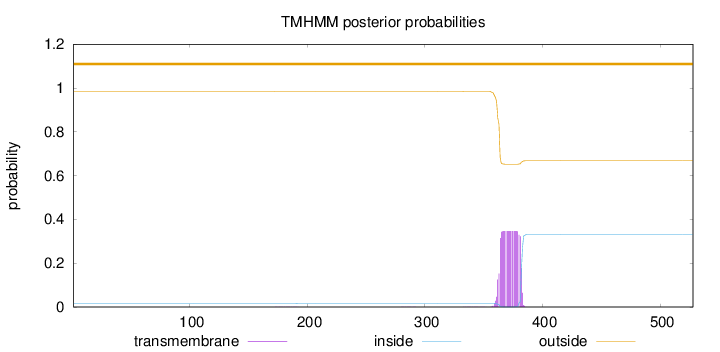
Topology
Subcellular location
Nucleus speckle
Length:
528
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.83066999999999
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.01517
outside
1 - 528
Population Genetic Test Statistics
Pi
203.436426
Theta
180.164304
Tajima's D
0.509973
CLR
0.014541
CSRT
0.516674166291685
Interpretation
Uncertain
