Gene
KWMTBOMO04473
Pre Gene Modal
BGIBMGA005437
Annotation
PREDICTED:_protein_spinster_isoform_X2_[Amyelois_transitella]
Full name
Protein spinster
Alternative Name
Protein benchwarmer
Protein diphthong
Protein diphthong
Location in the cell
PlasmaMembrane Reliability : 4.973
Sequence
CDS
ATGTCAATACAATATCAAGCAACAATCAGTGTTTTAAATAATGTCAAAACTGAATTCGAAATTGGTGATGGCAAGGCTGGATTTCTACAGACAGTGTTCGTCGTCGCGTACATGTTGTTCGCGCCAATATTTGGCTACTTAGGAGATAGATACTCAAGGCGCGTCATTATGGCCAGTGGCGTTGCGCTCTGGAGTCTAACAACTTTCTTTGGCTCGTTTTTACACGATTACGAGTCGTTTGCATTCTTCCGTGGTCTCGTTGGAATTGGAGAAGCCTCGTACTCCACGATAGCCCCTACCATCATCAGCGATCTCTTCGTGGGGGATGTAAGATCGAAGATGTTAGCATTCTTCTATTTTGCCATCCCAGTCGGAAGTGGTCTCGGATATATAGTAGGGTCAGTTGTGGGTGCAGCGCTAGGCAATTGGCGATACGGTCTACGGGTGACGCCTTTCTTTGGCGCGATTGCCGTCGCGCTCATCATATGGGTCATGGAAGACCCGGAACGAGGCGCCTCCGAAGAGAGTCACATAAAACCCACTTCGTATAAGGACGATCTGCGTGCTCTCATAAAAAATCCATCGTTCATGCTGTCGACGATCGCTTTTACATGCGTAGCGTTCGTGACCGGCGCGCTCACGTGGTGGGGACCGCAAGTTATTTACTTGGGTCTCGGCTTGCAACCTGGCAACGAGTTTTCCTTCGAAAGCGTGTCGTACAAGTTCGGGTTGGTGGGCATGGCGGCGGGCGCGCTGGGCGTGCCGCTGGGCTGCGCGGCGGCGGCGCGCGCGCGGCCCCTCGTGGCCGACGCCGACCCGCTGCTGTGCGGCGCCGCGCTGCTGCTCTCCGCGCCGCTCGTGTTCTGTGCGCTGATCGCCGTGCGTGCCTCCGCCGCGGGGACCTTCGCGCTCATCTTCTTCGGCATGCTCACTCTCAACCTCACCTGGTCCGTAGTCGCCGACATGATCCTGTACGTTGTCATACCACCAAGGCGTTCGACAGCAGAAGCTTTCCAGATATTAATATCGCACATGTTTGGGGATGCTGGAAGTCCGTACTTAGTCGGAGTTATATCAGAAAGTTTGAAGTCCTACTTGATGCCATCACCGGATGATTTGCCAAGTAAAACTGTAGAATTCAGAGCGCTTCAGTATGCGCTGTTCATAACGTGCTTCGTGGAAGTACTCGGAGGAATATTCTTCCTTATAACATCGGCGTACATTGTCAGGGACAAACTAAAAGTGGACAGAGCTATTGCAGAAGCAGAAGCACAGCACACGGAGCCGTCCCACGCTTCAGCACACGAAGACATTGTAGCCGGCGAATAG
Protein
MSIQYQATISVLNNVKTEFEIGDGKAGFLQTVFVVAYMLFAPIFGYLGDRYSRRVIMASGVALWSLTTFFGSFLHDYESFAFFRGLVGIGEASYSTIAPTIISDLFVGDVRSKMLAFFYFAIPVGSGLGYIVGSVVGAALGNWRYGLRVTPFFGAIAVALIIWVMEDPERGASEESHIKPTSYKDDLRALIKNPSFMLSTIAFTCVAFVTGALTWWGPQVIYLGLGLQPGNEFSFESVSYKFGLVGMAAGALGVPLGCAAAARARPLVADADPLLCGAALLLSAPLVFCALIAVRASAAGTFALIFFGMLTLNLTWSVVADMILYVVIPPRRSTAEAFQILISHMFGDAGSPYLVGVISESLKSYLMPSPDDLPSKTVEFRALQYALFITCFVEVLGGIFFLITSAYIVRDKLKVDRAIAEAEAQHTEPSHASAHEDIVAGE
Summary
Description
Probable sphingolipid transporter that plays a central role in endosomes and/or lysosomes storage. Involved in TGF-beta-mediated synaptic growth regulation both pre- and postsynaptically via its function in endosomal storage regulation. Also required during oogenesis by regulating yolk spheres storage.
Similarity
Belongs to the major facilitator superfamily. Spinster (TC 2.A.1.49) family.
Keywords
Alternative splicing
Complete proteome
Developmental protein
Differentiation
Endosome
Glycoprotein
Lipid transport
Lysosome
Membrane
Oogenesis
Reference proteome
Transmembrane
Transmembrane helix
Transport
Feature
chain Protein spinster
splice variant In isoform E.
splice variant In isoform E.
Uniprot
A0A2A4K150
H9J7E5
A0A2A4K019
A0A2A4JZG3
A0A2A4K0F0
A0A1Y1LAR1
+ More
A0A1Y1LAQ8 A0A1Y1LFH6 A0A1Y1LI80 A0A1B6F604 A0A336KI24 D6WWV4 A0A1L8DE93 A0A1B6IY79 A0A1B6JDS6 A0A336KDF3 A0A1B6JMB3 A0A336M1G7 A0A336MS93 A0A2J7RJL4 Q0IEQ4 A0A0K8TM89 A7UU76 A0A1I8JU77 A0A1Q3FWR3 A0A182K9W4 A0A2M3Z527 Q0IEQ3 Q7PIC0 A7UU77 A0A023F3L6 A0A0P9AGM4 A0A1Q3FXA7 A0A154PEF7 J9K505 A0A2M4BHV4 A0A0V0G8J6 A0A0P8XQY7 A0A2M4BH73 A0A0J9U3C6 A0A2S2QQ45 A0A2H8TVW2 N6V997 A0A0R1DST5 A0A2M4CLN4 A0A023EVL5 A0A2M4CLK5 A0A2S2PFH2 R4NR58 Q9GQQ0 A0A067R556 K4M4D5 A0A2M4A6N2 A0A2M4CLP9 A0A1B6JA87 A0A1Q3FX49 A0A0Q5VM11 A0A0Q9W6V7 A0A0J7N4Y2 A0A2Y9D1L0 A0A1B6HEE3 A0A0Q5VM10 A0A0J9RF33 A0A084WKA3 Q9GQQ0-4 W5JS35 A0A195BZK1 B4HSK2 A0A1J1HTU0 A0A158NP22 A0A3B0JL53 A0A1W4W4J4 Q9GQQ0-3 A0A1W4VBU4 B4P6Q9 A0A1J1HZD5 A0A1W4WF20 B3MF45 A0A0Q9WDE1 A0A0R3NQQ6 A0A1W4W4H5 A0A3Q0JGF3 U4U5S7 B4LL62 A0A151XDQ2 E2AXC6 B4H6B2 A0A1B0CTB9 Q28WT0 E9IEC1 A0A146M1V6 A0A3B0J088 B3NPX0 F4WGK1 A0A0L7QYX8 A0A0Q5W0Z4 Q9GQQ0-2 A0A195DXJ9 A0A026VZK9 A0A3B0JS91
A0A1Y1LAQ8 A0A1Y1LFH6 A0A1Y1LI80 A0A1B6F604 A0A336KI24 D6WWV4 A0A1L8DE93 A0A1B6IY79 A0A1B6JDS6 A0A336KDF3 A0A1B6JMB3 A0A336M1G7 A0A336MS93 A0A2J7RJL4 Q0IEQ4 A0A0K8TM89 A7UU76 A0A1I8JU77 A0A1Q3FWR3 A0A182K9W4 A0A2M3Z527 Q0IEQ3 Q7PIC0 A7UU77 A0A023F3L6 A0A0P9AGM4 A0A1Q3FXA7 A0A154PEF7 J9K505 A0A2M4BHV4 A0A0V0G8J6 A0A0P8XQY7 A0A2M4BH73 A0A0J9U3C6 A0A2S2QQ45 A0A2H8TVW2 N6V997 A0A0R1DST5 A0A2M4CLN4 A0A023EVL5 A0A2M4CLK5 A0A2S2PFH2 R4NR58 Q9GQQ0 A0A067R556 K4M4D5 A0A2M4A6N2 A0A2M4CLP9 A0A1B6JA87 A0A1Q3FX49 A0A0Q5VM11 A0A0Q9W6V7 A0A0J7N4Y2 A0A2Y9D1L0 A0A1B6HEE3 A0A0Q5VM10 A0A0J9RF33 A0A084WKA3 Q9GQQ0-4 W5JS35 A0A195BZK1 B4HSK2 A0A1J1HTU0 A0A158NP22 A0A3B0JL53 A0A1W4W4J4 Q9GQQ0-3 A0A1W4VBU4 B4P6Q9 A0A1J1HZD5 A0A1W4WF20 B3MF45 A0A0Q9WDE1 A0A0R3NQQ6 A0A1W4W4H5 A0A3Q0JGF3 U4U5S7 B4LL62 A0A151XDQ2 E2AXC6 B4H6B2 A0A1B0CTB9 Q28WT0 E9IEC1 A0A146M1V6 A0A3B0J088 B3NPX0 F4WGK1 A0A0L7QYX8 A0A0Q5W0Z4 Q9GQQ0-2 A0A195DXJ9 A0A026VZK9 A0A3B0JS91
Pubmed
19121390
28004739
18362917
19820115
17510324
26369729
+ More
12364791 14747013 17210077 25474469 17994087 22936249 15632085 17550304 24945155 11340170 10731132 12537572 12537569 12408844 15998804 24845553 24438588 20920257 23761445 21347285 23537049 20798317 21282665 26823975 21719571 24508170 30249741
12364791 14747013 17210077 25474469 17994087 22936249 15632085 17550304 24945155 11340170 10731132 12537572 12537569 12408844 15998804 24845553 24438588 20920257 23761445 21347285 23537049 20798317 21282665 26823975 21719571 24508170 30249741
EMBL
NWSH01000327
PCG77400.1
BABH01013680
BABH01013681
BABH01013682
BABH01013683
+ More
PCG77399.1 PCG77401.1 PCG77398.1 GEZM01061156 JAV70723.1 GEZM01061155 JAV70724.1 GEZM01061154 JAV70725.1 GEZM01061153 JAV70727.1 GECZ01024162 JAS45607.1 UFQS01000325 UFQT01000325 SSX02895.1 SSX23262.1 KQ971360 EFA09102.2 GFDF01009305 JAV04779.1 GECU01015815 JAS91891.1 GECU01010364 JAS97342.1 SSX02896.1 SSX23263.1 GECU01007315 JAT00392.1 UFQT01000391 SSX23890.1 UFQT01002419 SSX33414.1 NEVH01002992 PNF41022.1 CH477504 EAT39877.1 GDAI01002340 JAI15263.1 AAAB01008960 EDO63855.1 GFDL01002984 JAV32061.1 GGFM01002869 MBW23620.1 EAT39876.1 EAA44203.4 EDO63854.1 GBBI01002774 JAC15938.1 CH902619 KPU76967.1 GFDL01002963 JAV32082.1 KQ434874 KZC09658.1 ABLF02034683 GGFJ01003247 MBW52388.1 GECL01002338 JAP03786.1 KPU76968.1 GGFJ01003246 MBW52387.1 CM002911 KMY94180.1 GGMS01010648 MBY79851.1 GFXV01005877 MBW17682.1 CM000071 ENO01981.2 CM000158 KRK00232.1 GGFL01002015 MBW66193.1 GAPW01000517 JAC13081.1 GGFL01002016 MBW66194.1 GGMR01015570 MBY28189.1 BT150099 AGL46748.1 AF212366 AF212367 AF212368 AF212369 AF212370 AE013599 BT032842 AY051792 KK852687 KDR18407.1 BT149866 AFV15811.1 GGFK01002967 MBW36288.1 GGFL01002017 MBW66195.1 GECU01011609 JAS96097.1 GFDL01002989 JAV32056.1 CH954179 KQS62471.1 CH940648 KRF80294.1 LBMM01009983 KMQ87745.1 GECU01034660 JAS73046.1 KQS62470.1 KMY94179.1 ATLV01024104 KE525349 KFB50647.1 ADMH02000627 ETN65559.1 KQ976394 KYM93351.1 CH480816 EDW48080.1 CVRI01000021 CRK91488.1 ADTU01022127 ADTU01022128 OUUW01000001 SPP73966.1 EDW91986.2 CRK91489.1 EDV37673.2 KRF80295.1 KRT03196.1 KB631763 ERL85956.1 EDW61869.2 KQ982268 KYQ58504.1 GL443548 EFN61906.1 CH479213 EDW33336.1 AJWK01027427 EAL26587.2 GL762576 EFZ21086.1 GDHC01005220 JAQ13409.1 SPP73967.1 EDV55817.1 GL888137 EGI66666.1 KQ414685 KOC63820.1 KQS62472.1 KQ980167 KYN17339.1 KK107536 QOIP01000008 EZA49187.1 RLU19810.1 SPP73968.1
PCG77399.1 PCG77401.1 PCG77398.1 GEZM01061156 JAV70723.1 GEZM01061155 JAV70724.1 GEZM01061154 JAV70725.1 GEZM01061153 JAV70727.1 GECZ01024162 JAS45607.1 UFQS01000325 UFQT01000325 SSX02895.1 SSX23262.1 KQ971360 EFA09102.2 GFDF01009305 JAV04779.1 GECU01015815 JAS91891.1 GECU01010364 JAS97342.1 SSX02896.1 SSX23263.1 GECU01007315 JAT00392.1 UFQT01000391 SSX23890.1 UFQT01002419 SSX33414.1 NEVH01002992 PNF41022.1 CH477504 EAT39877.1 GDAI01002340 JAI15263.1 AAAB01008960 EDO63855.1 GFDL01002984 JAV32061.1 GGFM01002869 MBW23620.1 EAT39876.1 EAA44203.4 EDO63854.1 GBBI01002774 JAC15938.1 CH902619 KPU76967.1 GFDL01002963 JAV32082.1 KQ434874 KZC09658.1 ABLF02034683 GGFJ01003247 MBW52388.1 GECL01002338 JAP03786.1 KPU76968.1 GGFJ01003246 MBW52387.1 CM002911 KMY94180.1 GGMS01010648 MBY79851.1 GFXV01005877 MBW17682.1 CM000071 ENO01981.2 CM000158 KRK00232.1 GGFL01002015 MBW66193.1 GAPW01000517 JAC13081.1 GGFL01002016 MBW66194.1 GGMR01015570 MBY28189.1 BT150099 AGL46748.1 AF212366 AF212367 AF212368 AF212369 AF212370 AE013599 BT032842 AY051792 KK852687 KDR18407.1 BT149866 AFV15811.1 GGFK01002967 MBW36288.1 GGFL01002017 MBW66195.1 GECU01011609 JAS96097.1 GFDL01002989 JAV32056.1 CH954179 KQS62471.1 CH940648 KRF80294.1 LBMM01009983 KMQ87745.1 GECU01034660 JAS73046.1 KQS62470.1 KMY94179.1 ATLV01024104 KE525349 KFB50647.1 ADMH02000627 ETN65559.1 KQ976394 KYM93351.1 CH480816 EDW48080.1 CVRI01000021 CRK91488.1 ADTU01022127 ADTU01022128 OUUW01000001 SPP73966.1 EDW91986.2 CRK91489.1 EDV37673.2 KRF80295.1 KRT03196.1 KB631763 ERL85956.1 EDW61869.2 KQ982268 KYQ58504.1 GL443548 EFN61906.1 CH479213 EDW33336.1 AJWK01027427 EAL26587.2 GL762576 EFZ21086.1 GDHC01005220 JAQ13409.1 SPP73967.1 EDV55817.1 GL888137 EGI66666.1 KQ414685 KOC63820.1 KQS62472.1 KQ980167 KYN17339.1 KK107536 QOIP01000008 EZA49187.1 RLU19810.1 SPP73968.1
Proteomes
UP000218220
UP000005204
UP000007266
UP000235965
UP000008820
UP000007062
+ More
UP000075900 UP000075881 UP000007801 UP000076502 UP000007819 UP000001819 UP000002282 UP000000803 UP000027135 UP000008711 UP000008792 UP000036403 UP000075884 UP000030765 UP000000673 UP000078540 UP000001292 UP000183832 UP000005205 UP000268350 UP000192223 UP000192221 UP000079169 UP000030742 UP000075809 UP000000311 UP000008744 UP000092461 UP000007755 UP000053825 UP000078492 UP000053097 UP000279307
UP000075900 UP000075881 UP000007801 UP000076502 UP000007819 UP000001819 UP000002282 UP000000803 UP000027135 UP000008711 UP000008792 UP000036403 UP000075884 UP000030765 UP000000673 UP000078540 UP000001292 UP000183832 UP000005205 UP000268350 UP000192223 UP000192221 UP000079169 UP000030742 UP000075809 UP000000311 UP000008744 UP000092461 UP000007755 UP000053825 UP000078492 UP000053097 UP000279307
Pfam
PF07690 MFS_1
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2A4K150
H9J7E5
A0A2A4K019
A0A2A4JZG3
A0A2A4K0F0
A0A1Y1LAR1
+ More
A0A1Y1LAQ8 A0A1Y1LFH6 A0A1Y1LI80 A0A1B6F604 A0A336KI24 D6WWV4 A0A1L8DE93 A0A1B6IY79 A0A1B6JDS6 A0A336KDF3 A0A1B6JMB3 A0A336M1G7 A0A336MS93 A0A2J7RJL4 Q0IEQ4 A0A0K8TM89 A7UU76 A0A1I8JU77 A0A1Q3FWR3 A0A182K9W4 A0A2M3Z527 Q0IEQ3 Q7PIC0 A7UU77 A0A023F3L6 A0A0P9AGM4 A0A1Q3FXA7 A0A154PEF7 J9K505 A0A2M4BHV4 A0A0V0G8J6 A0A0P8XQY7 A0A2M4BH73 A0A0J9U3C6 A0A2S2QQ45 A0A2H8TVW2 N6V997 A0A0R1DST5 A0A2M4CLN4 A0A023EVL5 A0A2M4CLK5 A0A2S2PFH2 R4NR58 Q9GQQ0 A0A067R556 K4M4D5 A0A2M4A6N2 A0A2M4CLP9 A0A1B6JA87 A0A1Q3FX49 A0A0Q5VM11 A0A0Q9W6V7 A0A0J7N4Y2 A0A2Y9D1L0 A0A1B6HEE3 A0A0Q5VM10 A0A0J9RF33 A0A084WKA3 Q9GQQ0-4 W5JS35 A0A195BZK1 B4HSK2 A0A1J1HTU0 A0A158NP22 A0A3B0JL53 A0A1W4W4J4 Q9GQQ0-3 A0A1W4VBU4 B4P6Q9 A0A1J1HZD5 A0A1W4WF20 B3MF45 A0A0Q9WDE1 A0A0R3NQQ6 A0A1W4W4H5 A0A3Q0JGF3 U4U5S7 B4LL62 A0A151XDQ2 E2AXC6 B4H6B2 A0A1B0CTB9 Q28WT0 E9IEC1 A0A146M1V6 A0A3B0J088 B3NPX0 F4WGK1 A0A0L7QYX8 A0A0Q5W0Z4 Q9GQQ0-2 A0A195DXJ9 A0A026VZK9 A0A3B0JS91
A0A1Y1LAQ8 A0A1Y1LFH6 A0A1Y1LI80 A0A1B6F604 A0A336KI24 D6WWV4 A0A1L8DE93 A0A1B6IY79 A0A1B6JDS6 A0A336KDF3 A0A1B6JMB3 A0A336M1G7 A0A336MS93 A0A2J7RJL4 Q0IEQ4 A0A0K8TM89 A7UU76 A0A1I8JU77 A0A1Q3FWR3 A0A182K9W4 A0A2M3Z527 Q0IEQ3 Q7PIC0 A7UU77 A0A023F3L6 A0A0P9AGM4 A0A1Q3FXA7 A0A154PEF7 J9K505 A0A2M4BHV4 A0A0V0G8J6 A0A0P8XQY7 A0A2M4BH73 A0A0J9U3C6 A0A2S2QQ45 A0A2H8TVW2 N6V997 A0A0R1DST5 A0A2M4CLN4 A0A023EVL5 A0A2M4CLK5 A0A2S2PFH2 R4NR58 Q9GQQ0 A0A067R556 K4M4D5 A0A2M4A6N2 A0A2M4CLP9 A0A1B6JA87 A0A1Q3FX49 A0A0Q5VM11 A0A0Q9W6V7 A0A0J7N4Y2 A0A2Y9D1L0 A0A1B6HEE3 A0A0Q5VM10 A0A0J9RF33 A0A084WKA3 Q9GQQ0-4 W5JS35 A0A195BZK1 B4HSK2 A0A1J1HTU0 A0A158NP22 A0A3B0JL53 A0A1W4W4J4 Q9GQQ0-3 A0A1W4VBU4 B4P6Q9 A0A1J1HZD5 A0A1W4WF20 B3MF45 A0A0Q9WDE1 A0A0R3NQQ6 A0A1W4W4H5 A0A3Q0JGF3 U4U5S7 B4LL62 A0A151XDQ2 E2AXC6 B4H6B2 A0A1B0CTB9 Q28WT0 E9IEC1 A0A146M1V6 A0A3B0J088 B3NPX0 F4WGK1 A0A0L7QYX8 A0A0Q5W0Z4 Q9GQQ0-2 A0A195DXJ9 A0A026VZK9 A0A3B0JS91
PDB
3O65
E-value=0.0277269,
Score=88
Ontologies
GO
GO:0016021
GO:0055085
GO:0008643
GO:0090099
GO:0008582
GO:0045476
GO:0031902
GO:0005765
GO:0098793
GO:0048477
GO:0007040
GO:0009267
GO:0035193
GO:0008333
GO:0031982
GO:0045477
GO:0040011
GO:0048488
GO:0012501
GO:0006897
GO:0043067
GO:0006869
GO:0010001
GO:0045924
GO:0008347
GO:0007619
GO:0051124
GO:0005634
GO:0006260
GO:0006281
GO:0006310
GO:0003676
GO:0006418
GO:0009058
GO:0016742
GO:0003824
Topology
Subcellular location
Late endosome membrane
Lysosome membrane
Lysosome membrane
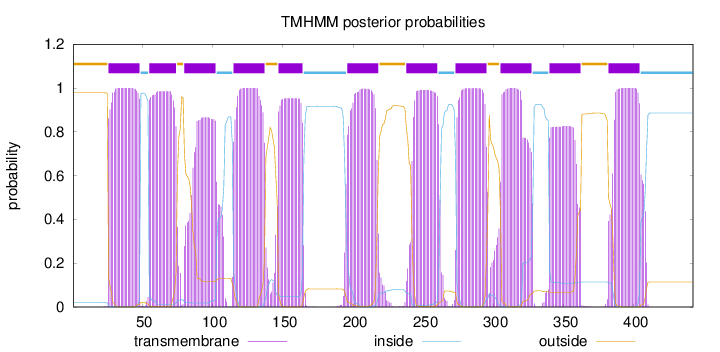
Length:
442
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
232.68414
Exp number, first 60 AAs:
28.45139
Total prob of N-in:
0.02179
POSSIBLE N-term signal
sequence
outside
1 - 25
TMhelix
26 - 48
inside
49 - 54
TMhelix
55 - 74
outside
75 - 79
TMhelix
80 - 102
inside
103 - 114
TMhelix
115 - 137
outside
138 - 146
TMhelix
147 - 164
inside
165 - 195
TMhelix
196 - 218
outside
219 - 237
TMhelix
238 - 260
inside
261 - 272
TMhelix
273 - 295
outside
296 - 304
TMhelix
305 - 327
inside
328 - 339
TMhelix
340 - 362
outside
363 - 381
TMhelix
382 - 404
inside
405 - 442
Population Genetic Test Statistics
Pi
373.209725
Theta
205.842541
Tajima's D
0.584506
CLR
0.142706
CSRT
0.537523123843808
Interpretation
Uncertain
