Gene
KWMTBOMO04467
Pre Gene Modal
BGIBMGA005373
Annotation
PREDICTED:_cationic_amino_acid_transporter_4_[Amyelois_transitella]
Full name
Lipoyl synthase, mitochondrial
Alternative Name
Lipoate synthase
Lipoic acid synthase
Lipoic acid synthase
Location in the cell
PlasmaMembrane Reliability : 4.946
Sequence
CDS
ATGCCGGGCGCAAGGCACAAGATCCTTGGCCATGTGCTATCCGGCTTCTGTCATAAAATGAATCGATGCAAGCCACTACACGGGGATTCCATGGATACTCCCCTAAATAGATGTCTAACCACTTTTGATATAACATTACTCGGCGTGGGTCACATGGTGGGTGCAGGGATATATGTACTTACCGGAACTGTTGCAAAATCAATGGCTGGTCCTTCGACTGCACTAAGTTTCCTTCTGGCGGGGATAACTTCGACTCTCGCTGCCCTTTGTTATGCTGAATTCGGGACTCGAATACCACGAGCGGGTAGCGCGTACGCTTATACCTATGTCAGTATTGGTGAATTTTGGGCCTTTATTATCGGATGGAATATTGTTTTGGAATATATGATTGGTGCTGCTTCAGTGGCGAGAGCTTGGTCAGGATATTTGGATTCTATATTACATGGGGCTATCAGTAATGCCACAGTAGCGTTAACTGGTGAACTTCATGAAACACTTTTAAGCAGATATCCTGATATTCTAGCGTTTCTCATATGTCTAGCAGCGTCCTTGATACTCGCTGCGGGTGTTAAAACATCAGCCTATATCAATAACGGCCTGACTATTTTAAATCTTTTGGTCATCTCTTTAGTAATCTTTTTGGGTTTTTATTATGCTGATATCAGCAACTGGTCAGAGAAAAATGGTGGTTTTATGCCGTTTGGTTTTAGTGGTGTGTTAGCAGGAGCTGCTACTTGTTTTTACGCTTTTGTAGGGTTTGATAGTATATCTGCTTCCAGTGAAGAAGCCAAAGATCCTTCTCGCTCTATTCCTGTTGCAACAGTATTATCAATGGCTGTAGTGGCTTTTGGTTATATTTTGGTCGCGATGGCATTAACTTTAATGGTTCCATATAATACAATTAATCCAGATGCTGCGCTTCCAGCTGCTCTTGGTGCAGTCCACGCAGACTGGGCAAAATATGCAGTTGCTGTGGGTGCAGTATGTGGAATGACAACAACTTTACTTGGATCCATGTTTTCACTACCACGATGTCTATATGCTATGTCTGCCGACGGTTTGCTTTTTGGGTTCTTCAGCGACATTAGTAACAAGGCCCAAATTCCTGTTGCTAATTTAGCTATTTCTGGTTTATCGTCTGCTTTCATAGCTCTTCTTTTTGATTTGGAAAAACTTGTAGAATTTATGTCTATAGGCACATTGTTAGCCTACACGATTGTTAGTGCCGCCGTAATAATATTGAGATATCGTCCAACTCCAACAACAGAAGATAAAGGTTTTGTTGTCCCACAGTTAGATTCACCTTGTGATCGAGAGGATAGTTCAGCAACTGGAACGCCGGCAACGGATGGCGGATCTTCTTCTTCTGAGATGTTTGAAACATTAACGGTCGGTCGTCTACGGCCTCAATTCGCATGGCTAGAACCTCTTGTTGGTGGTAGAGCCCCTGGTGCTGCTGTGACATGTTCTGTGTATACATTTACGATTGCTACTGCGGCTCTGTGTGCTCACAACCATTTTCTCGTCGAACCTGCTGGATTATGGGTGTTATTGCCGGACTTCGTTCTTATTTTCATCATCATCTCGTGTTTGGTGATAATCTGGGCTCACCAACAAAGTCCGACTCGTTTACCGTTCCGTGTACCTTGGGTACCACTGTTACCTGCTGCAAGTGTGATGCTCAACGTAGAACTTATGATTAATCTGAATGCTCTAACTTGGGCACGCTTTGCTATGTGGATGACTTTCGGTCTTCTTGTATATTTCCTATACGGTATCCATCACAGCAAGCTGGGAGAGGGTGTCGCCGGTTTGCTATCCAGCGGTGGAAATAACAGCGATTGGGGTGCAGTAGAAAAGACGAGTTCACGACGTATTGGCCGCTTTGGTCGGAGCAGTAAAGGAGATGACCGCAAACCCATCATTTGTGAAGACGAACTCTCTAGAAGAGAACCTTGA
Protein
MPGARHKILGHVLSGFCHKMNRCKPLHGDSMDTPLNRCLTTFDITLLGVGHMVGAGIYVLTGTVAKSMAGPSTALSFLLAGITSTLAALCYAEFGTRIPRAGSAYAYTYVSIGEFWAFIIGWNIVLEYMIGAASVARAWSGYLDSILHGAISNATVALTGELHETLLSRYPDILAFLICLAASLILAAGVKTSAYINNGLTILNLLVISLVIFLGFYYADISNWSEKNGGFMPFGFSGVLAGAATCFYAFVGFDSISASSEEAKDPSRSIPVATVLSMAVVAFGYILVAMALTLMVPYNTINPDAALPAALGAVHADWAKYAVAVGAVCGMTTTLLGSMFSLPRCLYAMSADGLLFGFFSDISNKAQIPVANLAISGLSSAFIALLFDLEKLVEFMSIGTLLAYTIVSAAVIILRYRPTPTTEDKGFVVPQLDSPCDREDSSATGTPATDGGSSSSEMFETLTVGRLRPQFAWLEPLVGGRAPGAAVTCSVYTFTIATAALCAHNHFLVEPAGLWVLLPDFVLIFIIISCLVIIWAHQQSPTRLPFRVPWVPLLPAASVMLNVELMINLNALTWARFAMWMTFGLLVYFLYGIHHSKLGEGVAGLLSSGGNNSDWGAVEKTSSRRIGRFGRSSKGDDRKPIICEDELSRREP
Summary
Description
Catalyzes the radical-mediated insertion of two sulfur atoms into the C-6 and C-8 positions of the octanoyl moiety bound to the lipoyl domains of lipoate-dependent enzymes, thereby converting the octanoylated domains into lipoylated derivatives.
Catalytic Activity
[[Fe-S] cluster scaffold protein carrying a second [4Fe-4S](2+) cluster] + 4 H(+) + N(6)-octanoyl-L-lysyl-[protein] + 2 oxidized [2Fe-2S]-[ferredoxin] + 2 S-adenosyl-L-methionine = (R)-N(6)-dihydrolipoyl-L-lysyl-[protein] + 2 5'-deoxyadenosine + [[Fe-S] cluster scaffold protein] + 4 Fe(3+) + 2 hydrogen sulfide + 2 L-methionine + 2 reduced [2Fe-2S]-[ferredoxin]
Cofactor
[4Fe-4S] cluster
Similarity
Belongs to the radical SAM superfamily. Lipoyl synthase family.
Uniprot
H9J781
A0A2H1W7F3
A0A2A4JZH1
A0A212FEP9
A0A194PM86
A0A194QSG4
+ More
A0A3S2NI70 A0A0L7LAT9 A0A1B6D6M1 A0A067QSN3 A0A2R7WD22 A0A2J7PHM4 T1I0G7 A0A023F3A9 A0A084VKX1 A0A182JRK1 B0WAU5 A0A182MYD5 A0A182QTQ2 Q7Q8T5 B4KYY5 A0A182JKW0 A0A182LQ24 Q16W21 B4J3M8 A0A182X1V2 A0A182HFR6 A0A182YA31 A0A182P8H5 A0A0M3QWS7 W5JEE3 A0A182G9I6 A0A182S4V6 A0A182SPN1 A0A182URT1 A0A0L0C7Y6 A0A182FUK1 A0A2H8TCN8 A0A336MAD2 A0A1W4WIX7 Q2LYJ7 B4HBV1 A0A3B0K3T6 B4LD69 A0A2A3E9C6 B3M989 B3NIL2 A0A1W4W6S6 V9IDC4 A0A1B0GHL3 Q9VPG2 B4PF89 B4QRY6 Q6NNV9 D6WY56 B4MLJ1 A0A154PG88 K7IU03 A0A0L7R6N6 A0A151WP18 F4X1B7 A0A195DP28 A0A232EP67 A0A151JSU7 A0A164QTX2 A0A0P5SZP7 A0A0N8E7Z0 A0A0N0BKD7 A0A2P8YWU4 A0A182VTB7 Q16N09 A0A195BCN1 A0A088ASR1 A0A1B0DAS8 A0A0P6A948 A0A182U709 A0A0C9R8E4 A0A182MCA8 A0A0P6JV95 A0A1I8N3M4 E2B0W3 A0A1I8PJB1 A0A0T6B709 A0A3L8DAR3 A0A026WWI0 T1IVD2 A0A3B0K1Z5 V9IFJ9 A0A195D727 A0A3Q0J2D8 A0A158NFQ8 A0A2M3Z5B4 A0A2M3Z5F6 A0A0P5B888 A0A0P5W918 A0A0N8A1V7 W5KCS5 A0A1W4YFI1 A0A2D0QXU2 F6SWF3
A0A3S2NI70 A0A0L7LAT9 A0A1B6D6M1 A0A067QSN3 A0A2R7WD22 A0A2J7PHM4 T1I0G7 A0A023F3A9 A0A084VKX1 A0A182JRK1 B0WAU5 A0A182MYD5 A0A182QTQ2 Q7Q8T5 B4KYY5 A0A182JKW0 A0A182LQ24 Q16W21 B4J3M8 A0A182X1V2 A0A182HFR6 A0A182YA31 A0A182P8H5 A0A0M3QWS7 W5JEE3 A0A182G9I6 A0A182S4V6 A0A182SPN1 A0A182URT1 A0A0L0C7Y6 A0A182FUK1 A0A2H8TCN8 A0A336MAD2 A0A1W4WIX7 Q2LYJ7 B4HBV1 A0A3B0K3T6 B4LD69 A0A2A3E9C6 B3M989 B3NIL2 A0A1W4W6S6 V9IDC4 A0A1B0GHL3 Q9VPG2 B4PF89 B4QRY6 Q6NNV9 D6WY56 B4MLJ1 A0A154PG88 K7IU03 A0A0L7R6N6 A0A151WP18 F4X1B7 A0A195DP28 A0A232EP67 A0A151JSU7 A0A164QTX2 A0A0P5SZP7 A0A0N8E7Z0 A0A0N0BKD7 A0A2P8YWU4 A0A182VTB7 Q16N09 A0A195BCN1 A0A088ASR1 A0A1B0DAS8 A0A0P6A948 A0A182U709 A0A0C9R8E4 A0A182MCA8 A0A0P6JV95 A0A1I8N3M4 E2B0W3 A0A1I8PJB1 A0A0T6B709 A0A3L8DAR3 A0A026WWI0 T1IVD2 A0A3B0K1Z5 V9IFJ9 A0A195D727 A0A3Q0J2D8 A0A158NFQ8 A0A2M3Z5B4 A0A2M3Z5F6 A0A0P5B888 A0A0P5W918 A0A0N8A1V7 W5KCS5 A0A1W4YFI1 A0A2D0QXU2 F6SWF3
EC Number
2.8.1.8
Pubmed
19121390
22118469
26354079
26227816
24845553
25474469
+ More
24438588 12364791 17994087 20966253 17510324 25244985 20920257 23761445 26483478 26108605 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 18362917 19820115 20075255 21719571 28648823 29403074 25315136 20798317 30249741 24508170 21347285 25329095 20431018
24438588 12364791 17994087 20966253 17510324 25244985 20920257 23761445 26483478 26108605 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 18362917 19820115 20075255 21719571 28648823 29403074 25315136 20798317 30249741 24508170 21347285 25329095 20431018
EMBL
BABH01013671
BABH01013672
BABH01013673
ODYU01006801
SOQ48988.1
NWSH01000327
+ More
PCG77415.1 AGBW02008899 OWR52235.1 KQ459599 KPI94437.1 KQ461154 KPJ08427.1 RSAL01000088 RVE48220.1 JTDY01001897 KOB72592.1 GEDC01015975 JAS21323.1 KK853329 KDR08430.1 KK854586 PTY17141.1 NEVH01025135 PNF15825.1 ACPB03016100 GBBI01002812 JAC15900.1 ATLV01014310 KE524956 KFB38615.1 DS231875 EDS41811.1 AXCN02002053 AAAB01008933 EAA09935.2 CH933809 EDW18877.1 CH477577 EAT38775.1 CH916366 EDV97259.1 APCN01005567 CP012525 ALC44629.1 ADMH02001479 ETN62406.1 JXUM01049229 JXUM01049230 KQ561596 KXJ78053.1 JRES01000760 KNC28543.1 GFXV01000072 MBW11877.1 UFQT01000621 SSX25849.1 CH379069 EAL29865.3 CH479267 EDW39545.1 OUUW01000012 SPP87342.1 CH940647 EDW69950.1 KZ288334 PBC27882.1 CH902618 EDV40073.2 CH954178 EDV52508.1 JR040284 AEY59065.1 AJWK01005316 AE014296 BT099767 AAF51591.1 ACV82466.1 CM000159 EDW95175.1 CM000363 CM002912 EDX11232.1 KMZ00780.1 BT011174 AAR88535.1 KQ971362 EFA08956.2 CH963847 EDW72847.1 KQ434890 KZC10464.1 KQ414646 KOC66411.1 KQ982893 KYQ49558.1 GL888528 EGI59789.1 KQ980662 KYN14670.1 NNAY01002993 OXU20155.1 KQ982011 KYN30675.1 LRGB01002371 KZS08057.1 GDIP01132985 JAL70729.1 GDIQ01156015 GDIQ01153355 GDIQ01052948 JAN41789.1 KQ435700 KOX80454.1 PYGN01000312 PSN48720.1 CH477843 EAT35719.1 KQ976514 KYM82313.1 AJVK01013390 AJVK01013391 GDIP01033145 JAM70570.1 GBYB01004310 JAG74077.1 AXCM01009715 GDIQ01007481 JAN87256.1 GL444701 EFN60666.1 LJIG01009402 KRT83151.1 QOIP01000010 RLU17401.1 KK107079 EZA60168.1 JH431581 SPP87343.1 JR040283 AEY59064.1 KQ976750 KYN08681.1 ADTU01014565 ADTU01014566 GGFM01002952 MBW23703.1 GGFM01002983 MBW23734.1 GDIP01188085 JAJ35317.1 GDIP01089504 JAM14211.1 GDIP01190681 JAJ32721.1 AAMC01008374
PCG77415.1 AGBW02008899 OWR52235.1 KQ459599 KPI94437.1 KQ461154 KPJ08427.1 RSAL01000088 RVE48220.1 JTDY01001897 KOB72592.1 GEDC01015975 JAS21323.1 KK853329 KDR08430.1 KK854586 PTY17141.1 NEVH01025135 PNF15825.1 ACPB03016100 GBBI01002812 JAC15900.1 ATLV01014310 KE524956 KFB38615.1 DS231875 EDS41811.1 AXCN02002053 AAAB01008933 EAA09935.2 CH933809 EDW18877.1 CH477577 EAT38775.1 CH916366 EDV97259.1 APCN01005567 CP012525 ALC44629.1 ADMH02001479 ETN62406.1 JXUM01049229 JXUM01049230 KQ561596 KXJ78053.1 JRES01000760 KNC28543.1 GFXV01000072 MBW11877.1 UFQT01000621 SSX25849.1 CH379069 EAL29865.3 CH479267 EDW39545.1 OUUW01000012 SPP87342.1 CH940647 EDW69950.1 KZ288334 PBC27882.1 CH902618 EDV40073.2 CH954178 EDV52508.1 JR040284 AEY59065.1 AJWK01005316 AE014296 BT099767 AAF51591.1 ACV82466.1 CM000159 EDW95175.1 CM000363 CM002912 EDX11232.1 KMZ00780.1 BT011174 AAR88535.1 KQ971362 EFA08956.2 CH963847 EDW72847.1 KQ434890 KZC10464.1 KQ414646 KOC66411.1 KQ982893 KYQ49558.1 GL888528 EGI59789.1 KQ980662 KYN14670.1 NNAY01002993 OXU20155.1 KQ982011 KYN30675.1 LRGB01002371 KZS08057.1 GDIP01132985 JAL70729.1 GDIQ01156015 GDIQ01153355 GDIQ01052948 JAN41789.1 KQ435700 KOX80454.1 PYGN01000312 PSN48720.1 CH477843 EAT35719.1 KQ976514 KYM82313.1 AJVK01013390 AJVK01013391 GDIP01033145 JAM70570.1 GBYB01004310 JAG74077.1 AXCM01009715 GDIQ01007481 JAN87256.1 GL444701 EFN60666.1 LJIG01009402 KRT83151.1 QOIP01000010 RLU17401.1 KK107079 EZA60168.1 JH431581 SPP87343.1 JR040283 AEY59064.1 KQ976750 KYN08681.1 ADTU01014565 ADTU01014566 GGFM01002952 MBW23703.1 GGFM01002983 MBW23734.1 GDIP01188085 JAJ35317.1 GDIP01089504 JAM14211.1 GDIP01190681 JAJ32721.1 AAMC01008374
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000283053
+ More
UP000037510 UP000027135 UP000235965 UP000015103 UP000030765 UP000075881 UP000002320 UP000075884 UP000075886 UP000007062 UP000009192 UP000075880 UP000075882 UP000008820 UP000001070 UP000076407 UP000075840 UP000076408 UP000075885 UP000092553 UP000000673 UP000069940 UP000249989 UP000075900 UP000075901 UP000075903 UP000037069 UP000069272 UP000192223 UP000001819 UP000008744 UP000268350 UP000008792 UP000242457 UP000007801 UP000008711 UP000192221 UP000092461 UP000000803 UP000002282 UP000000304 UP000007266 UP000007798 UP000076502 UP000002358 UP000053825 UP000075809 UP000007755 UP000078492 UP000215335 UP000078541 UP000076858 UP000053105 UP000245037 UP000075920 UP000078540 UP000005203 UP000092462 UP000075902 UP000075883 UP000095301 UP000000311 UP000095300 UP000279307 UP000053097 UP000078542 UP000079169 UP000005205 UP000018467 UP000192224 UP000221080 UP000008143
UP000037510 UP000027135 UP000235965 UP000015103 UP000030765 UP000075881 UP000002320 UP000075884 UP000075886 UP000007062 UP000009192 UP000075880 UP000075882 UP000008820 UP000001070 UP000076407 UP000075840 UP000076408 UP000075885 UP000092553 UP000000673 UP000069940 UP000249989 UP000075900 UP000075901 UP000075903 UP000037069 UP000069272 UP000192223 UP000001819 UP000008744 UP000268350 UP000008792 UP000242457 UP000007801 UP000008711 UP000192221 UP000092461 UP000000803 UP000002282 UP000000304 UP000007266 UP000007798 UP000076502 UP000002358 UP000053825 UP000075809 UP000007755 UP000078492 UP000215335 UP000078541 UP000076858 UP000053105 UP000245037 UP000075920 UP000078540 UP000005203 UP000092462 UP000075902 UP000075883 UP000095301 UP000000311 UP000095300 UP000279307 UP000053097 UP000078542 UP000079169 UP000005205 UP000018467 UP000192224 UP000221080 UP000008143
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9J781
A0A2H1W7F3
A0A2A4JZH1
A0A212FEP9
A0A194PM86
A0A194QSG4
+ More
A0A3S2NI70 A0A0L7LAT9 A0A1B6D6M1 A0A067QSN3 A0A2R7WD22 A0A2J7PHM4 T1I0G7 A0A023F3A9 A0A084VKX1 A0A182JRK1 B0WAU5 A0A182MYD5 A0A182QTQ2 Q7Q8T5 B4KYY5 A0A182JKW0 A0A182LQ24 Q16W21 B4J3M8 A0A182X1V2 A0A182HFR6 A0A182YA31 A0A182P8H5 A0A0M3QWS7 W5JEE3 A0A182G9I6 A0A182S4V6 A0A182SPN1 A0A182URT1 A0A0L0C7Y6 A0A182FUK1 A0A2H8TCN8 A0A336MAD2 A0A1W4WIX7 Q2LYJ7 B4HBV1 A0A3B0K3T6 B4LD69 A0A2A3E9C6 B3M989 B3NIL2 A0A1W4W6S6 V9IDC4 A0A1B0GHL3 Q9VPG2 B4PF89 B4QRY6 Q6NNV9 D6WY56 B4MLJ1 A0A154PG88 K7IU03 A0A0L7R6N6 A0A151WP18 F4X1B7 A0A195DP28 A0A232EP67 A0A151JSU7 A0A164QTX2 A0A0P5SZP7 A0A0N8E7Z0 A0A0N0BKD7 A0A2P8YWU4 A0A182VTB7 Q16N09 A0A195BCN1 A0A088ASR1 A0A1B0DAS8 A0A0P6A948 A0A182U709 A0A0C9R8E4 A0A182MCA8 A0A0P6JV95 A0A1I8N3M4 E2B0W3 A0A1I8PJB1 A0A0T6B709 A0A3L8DAR3 A0A026WWI0 T1IVD2 A0A3B0K1Z5 V9IFJ9 A0A195D727 A0A3Q0J2D8 A0A158NFQ8 A0A2M3Z5B4 A0A2M3Z5F6 A0A0P5B888 A0A0P5W918 A0A0N8A1V7 W5KCS5 A0A1W4YFI1 A0A2D0QXU2 F6SWF3
A0A3S2NI70 A0A0L7LAT9 A0A1B6D6M1 A0A067QSN3 A0A2R7WD22 A0A2J7PHM4 T1I0G7 A0A023F3A9 A0A084VKX1 A0A182JRK1 B0WAU5 A0A182MYD5 A0A182QTQ2 Q7Q8T5 B4KYY5 A0A182JKW0 A0A182LQ24 Q16W21 B4J3M8 A0A182X1V2 A0A182HFR6 A0A182YA31 A0A182P8H5 A0A0M3QWS7 W5JEE3 A0A182G9I6 A0A182S4V6 A0A182SPN1 A0A182URT1 A0A0L0C7Y6 A0A182FUK1 A0A2H8TCN8 A0A336MAD2 A0A1W4WIX7 Q2LYJ7 B4HBV1 A0A3B0K3T6 B4LD69 A0A2A3E9C6 B3M989 B3NIL2 A0A1W4W6S6 V9IDC4 A0A1B0GHL3 Q9VPG2 B4PF89 B4QRY6 Q6NNV9 D6WY56 B4MLJ1 A0A154PG88 K7IU03 A0A0L7R6N6 A0A151WP18 F4X1B7 A0A195DP28 A0A232EP67 A0A151JSU7 A0A164QTX2 A0A0P5SZP7 A0A0N8E7Z0 A0A0N0BKD7 A0A2P8YWU4 A0A182VTB7 Q16N09 A0A195BCN1 A0A088ASR1 A0A1B0DAS8 A0A0P6A948 A0A182U709 A0A0C9R8E4 A0A182MCA8 A0A0P6JV95 A0A1I8N3M4 E2B0W3 A0A1I8PJB1 A0A0T6B709 A0A3L8DAR3 A0A026WWI0 T1IVD2 A0A3B0K1Z5 V9IFJ9 A0A195D727 A0A3Q0J2D8 A0A158NFQ8 A0A2M3Z5B4 A0A2M3Z5F6 A0A0P5B888 A0A0P5W918 A0A0N8A1V7 W5KCS5 A0A1W4YFI1 A0A2D0QXU2 F6SWF3
PDB
6F34
E-value=1.33463e-64,
Score=627
Ontologies
GO
Topology
Subcellular location
Mitochondrion
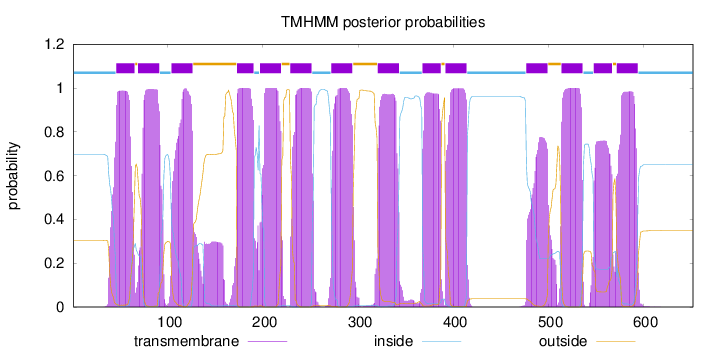
Length:
652
Number of predicted TMHs:
14
Exp number of AAs in TMHs:
297.93206
Exp number, first 60 AAs:
18.40728
Total prob of N-in:
0.69655
POSSIBLE N-term signal
sequence
inside
1 - 45
TMhelix
46 - 65
outside
66 - 68
TMhelix
69 - 91
inside
92 - 103
TMhelix
104 - 126
outside
127 - 172
TMhelix
173 - 190
inside
191 - 196
TMhelix
197 - 219
outside
220 - 228
TMhelix
229 - 251
inside
252 - 271
TMhelix
272 - 294
outside
295 - 320
TMhelix
321 - 343
inside
344 - 367
TMhelix
368 - 387
outside
388 - 391
TMhelix
392 - 414
inside
415 - 476
TMhelix
477 - 499
outside
500 - 513
TMhelix
514 - 536
inside
537 - 547
TMhelix
548 - 567
outside
568 - 571
TMhelix
572 - 594
inside
595 - 652
Population Genetic Test Statistics
Pi
208.586727
Theta
210.558069
Tajima's D
0.023265
CLR
0.720468
CSRT
0.375331233438328
Interpretation
Uncertain
