Gene
KWMTBOMO04465
Pre Gene Modal
BGIBMGA005374
Annotation
PREDICTED:_uncharacterized_protein_F21D5.5_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 2.378
Sequence
CDS
ATGTCTCGTTTATGCTTTTTGAGATGTTTGTTAGATACACATGCGCCTATTAAACTTCCTCACAACGTACAAATAATTGTAGGGCGCAATAAAGAAACAAAAATTAAGGATCAGTCATGTTCTAGACAGCAGCTAAGCTTAAAAGCAGATTGTGAAAAATGTCATGTAGAGATAAAACAACTAGGGGTAAACGCTTCTGGGTTAGATGGTTTTGCTCTTAAAAAAGATGAAGTGTATGAAATTGGACATGGATCAACAATTGAGATTTTATTAAACAATCATGTGCATATACTTGAATTTGATCCACCTCCTGAAAGCCTAACTACCTCACAGAATCAGACATTCAAAAGGAAATTGGAAGATGACAGTACATCTCAGAGAAAGAAATCATTAAAAGTGGAACAAGTGTCATCAAGTGTAAAAGAAAAAGTTGAAAAAGTTATTAGAGATATGTGGGATGAAATAGACAAAGGTGAATTGTATGTGTTTACAGCTAAGGGAGTGAAATCTAGTGACAAAATAGCATCTTTTGATATGGATGGAACCCTAATCAAGACAAAGTCTGGTAAAGTTCATCCGGTGGACATAAATGACTGGCAAATAGCATTTCCTTCAGTGCCCCAAAAACTGAAAGAATTTTTAGAAAAAGATTACAAAATTGTGTTATTTAGCAATCAAGCCCCTATTGGTAATGGTAGAGTTAAAATAGAAGACTTTAAAAAGAAGATTGAAAGTATTGTCCAAAAATTAAATGTGCCAGTACAAGCATATATAGCAACTGGTAAAGGGATTTACAGGAAACCTAGAATTGGAATGTGGATGACTTTGACTGAAAAGAAAAATGATGGTGTCAAAGTTGATATGAACCAAAGTTTTTATTGTGGAGATGCTGCAGGAAGGGTGGCAAACTGGGCCCCTGGTAAAAAGAAAGATCACTCCTTGGCAGACAAACTCTTTGCTGAAAATTTGGAATTAAAATTTTATACTCCCGAACAATTCTTCTTGGGACATTCTATTGCTAATGTGCTAATGAGTAAACCTGAGTTTCATCCTAAAGAAGTAACTGTAGAACCTTTTAATAATAAACTAATTAGCAAAGATAAAGAGATACTAATTTTAGTTGGATTTCCTGGTAGTGGAAAATCTTATTTGGCAAAGCAGATTGAAACTAAATCAAATCACAAATATGCGACTGTGTGCAGAGATATTCTAGGCTCATGGCAAAAATGTGCTGCTGAAGCCACAAAATTGTTGCAGCAAGGAAAAAGTGTCATAGTAGACAGTACGAATCCGGATGTTGAGTCTCGTTCCCGTTGGACATCACTCGCAAAACAAAATAAAGTAGATTGTCGTTGTGCTAAAATGGCTACTTCTAAGGCCCATGCGCAACATAATAACAAATTTAGGGAACTGATGAAGATCAATCATGTTCCTGTTAACGATATTGTATTCCATACCTACAAAAACAAGTTCACTGAACCGTCACCAAACGAAGGTTTTAAAGAAGTAATTGAAGTGAAATTCAACCCGTGCTTTGAGAGCGAAGAAGCTGAAAATTTATATAGAATGCATTTGTTAGAGAAATAA
Protein
MSRLCFLRCLLDTHAPIKLPHNVQIIVGRNKETKIKDQSCSRQQLSLKADCEKCHVEIKQLGVNASGLDGFALKKDEVYEIGHGSTIEILLNNHVHILEFDPPPESLTTSQNQTFKRKLEDDSTSQRKKSLKVEQVSSSVKEKVEKVIRDMWDEIDKGELYVFTAKGVKSSDKIASFDMDGTLIKTKSGKVHPVDINDWQIAFPSVPQKLKEFLEKDYKIVLFSNQAPIGNGRVKIEDFKKKIESIVQKLNVPVQAYIATGKGIYRKPRIGMWMTLTEKKNDGVKVDMNQSFYCGDAAGRVANWAPGKKKDHSLADKLFAENLELKFYTPEQFFLGHSIANVLMSKPEFHPKEVTVEPFNNKLISKDKEILILVGFPGSGKSYLAKQIETKSNHKYATVCRDILGSWQKCAAEATKLLQQGKSVIVDSTNPDVESRSRWTSLAKQNKVDCRCAKMATSKAHAQHNNKFRELMKINHVPVNDIVFHTYKNKFTEPSPNEGFKEVIEVKFNPCFESEEAENLYRMHLLEK
Summary
Uniprot
H9J782
A0A194QSY1
A0A194PTH4
A0A2A4K194
A0A2H1W7D6
A0A212FEU4
+ More
A0A2A4K0X3 A0A0L7L2G9 S4PXZ7 A0A067R0U4 A0A2J7RIJ5 E9G6T6 A0A0P5FHV9 A0A0P4X2H9 A0A151ILC0 A0A0N0BJP1 A0A026X339 Q17NZ7 D6WPR6 A0A182MXB4 A0A182MVD1 A0A182WDA0 A0A1B6H0S1 W5JNH4 E2C202 A0A195FVI1 A0A1S4EW42 A0A084WUK1 A0A182J3G3 A0A1B0D571 A0A158NLS9 A0A182JRT7 A0A182RAJ5 A0A195BLQ4 A0A0P4WQL4 E9J381 U5EV32 A0A1B6MDR6 K7IRQ0 A0A0L7QV67 A0A2J7RIJ1 A0A087ZW86 A0A1W4WYC1 E0VFJ9 A0A2A3EMX4 A0A154PSF1 A0A3R7LYR5 A0A182YQ91 A0A1B0C960 A0A2D4HFE4 A0A310SAT2 A0A336MJT2 A0A232FBV7 G1KMT2 A0A182PGC7 A0A1Y1KGB3 V4A8X4 A0A0C9R1P0 A0A0M4ENH1 A0A1B6FCW3 A0A336MNP9 W8C5W1 B8ZX67 A0A3B0JJY0 A0A1U7R7V0 A0A182QB07 B4K8T9 A0A182FTM2 B0WU84 A0A151NJZ6 A0A0F7ZAJ2 B4PT50 A0A1Q3F2C9 A0A0C9QEX3 A0A2L2Y9C8 R7V767 A0A2L2Y9D1 H0WRT3 A0A2K6JZR0 A0A2K6RGL1 A0A2R8ZCX7 K7BAU7 A0A0L0CLW8 J9JQT8 B3M0E7 B4LYW2 A0A0C9QMN2 A0A0P5V0A1 A0A0N7ZSL4 A0A1B0B4Z3 A0A0P5TWY5 A0A034W8B2 A0A2R8M3W8 A0A2U9B5U3 H9EWT0 F6PK17 A0A1A9ZT26 B4G3Q7 H9YYQ1
A0A2A4K0X3 A0A0L7L2G9 S4PXZ7 A0A067R0U4 A0A2J7RIJ5 E9G6T6 A0A0P5FHV9 A0A0P4X2H9 A0A151ILC0 A0A0N0BJP1 A0A026X339 Q17NZ7 D6WPR6 A0A182MXB4 A0A182MVD1 A0A182WDA0 A0A1B6H0S1 W5JNH4 E2C202 A0A195FVI1 A0A1S4EW42 A0A084WUK1 A0A182J3G3 A0A1B0D571 A0A158NLS9 A0A182JRT7 A0A182RAJ5 A0A195BLQ4 A0A0P4WQL4 E9J381 U5EV32 A0A1B6MDR6 K7IRQ0 A0A0L7QV67 A0A2J7RIJ1 A0A087ZW86 A0A1W4WYC1 E0VFJ9 A0A2A3EMX4 A0A154PSF1 A0A3R7LYR5 A0A182YQ91 A0A1B0C960 A0A2D4HFE4 A0A310SAT2 A0A336MJT2 A0A232FBV7 G1KMT2 A0A182PGC7 A0A1Y1KGB3 V4A8X4 A0A0C9R1P0 A0A0M4ENH1 A0A1B6FCW3 A0A336MNP9 W8C5W1 B8ZX67 A0A3B0JJY0 A0A1U7R7V0 A0A182QB07 B4K8T9 A0A182FTM2 B0WU84 A0A151NJZ6 A0A0F7ZAJ2 B4PT50 A0A1Q3F2C9 A0A0C9QEX3 A0A2L2Y9C8 R7V767 A0A2L2Y9D1 H0WRT3 A0A2K6JZR0 A0A2K6RGL1 A0A2R8ZCX7 K7BAU7 A0A0L0CLW8 J9JQT8 B3M0E7 B4LYW2 A0A0C9QMN2 A0A0P5V0A1 A0A0N7ZSL4 A0A1B0B4Z3 A0A0P5TWY5 A0A034W8B2 A0A2R8M3W8 A0A2U9B5U3 H9EWT0 F6PK17 A0A1A9ZT26 B4G3Q7 H9YYQ1
Pubmed
19121390
26354079
22118469
26227816
23622113
24845553
+ More
21292972 24508170 17510324 18362917 19820115 20920257 23761445 20798317 24438588 21347285 21282665 20075255 20566863 25244985 28648823 28004739 23254933 24495485 17994087 22293439 17550304 26561354 25362486 22722832 16136131 26108605 25348373 25319552 17431167
21292972 24508170 17510324 18362917 19820115 20920257 23761445 20798317 24438588 21347285 21282665 20075255 20566863 25244985 28648823 28004739 23254933 24495485 17994087 22293439 17550304 26561354 25362486 22722832 16136131 26108605 25348373 25319552 17431167
EMBL
BABH01013668
BABH01013669
KQ461154
KPJ08429.1
KQ459599
KPI94435.1
+ More
NWSH01000327 PCG77412.1 ODYU01006801 SOQ48990.1 AGBW02008899 OWR52233.1 PCG77413.1 JTDY01003451 KOB69536.1 GAIX01004168 JAA88392.1 KK852796 KDR16388.1 NEVH01003500 PNF40654.1 GL732533 EFX85136.1 GDIQ01257769 JAJ93955.1 GDIP01247258 LRGB01001579 JAI76143.1 KZS11549.1 KQ977121 KYN05544.1 KQ435713 KOX79322.1 KK107019 EZA62695.1 CH477195 EAT48401.1 KQ971354 EFA06186.1 AXCM01011152 GECZ01001504 JAS68265.1 ADMH02001047 ETN64329.1 GL452030 EFN78039.1 KQ981219 KYN44468.1 ATLV01027095 KE525423 KFB53895.1 AXCP01008618 AJVK01003341 ADTU01019796 KQ976450 KYM85732.1 GDRN01048555 JAI66647.1 GL768059 EFZ12716.1 GANO01001199 JAB58672.1 GEBQ01005948 JAT34029.1 KQ414727 KOC62530.1 PNF40655.1 AAZO01001857 DS235114 EEB12155.1 KZ288215 PBC32499.1 KQ435127 KZC14846.1 QCYY01002878 ROT66903.1 AJWK01001947 IACK01027967 LAA70675.1 KQ776210 OAD52211.1 UFQT01001048 SSX28617.1 NNAY01000508 OXU27943.1 GEZM01087528 JAV58645.1 KB202544 ESO89756.1 GBYB01001890 JAG71657.1 CP012526 ALC45460.1 GECZ01021734 JAS48035.1 UFQT01001411 SSX30433.1 GAMC01004649 JAC01907.1 FM956649 CAX11689.1 OUUW01000007 SPP82657.1 AXCN02001681 CH933806 EDW15508.1 DS232102 EDS34855.1 AKHW03002822 KYO37124.1 GBEX01000375 JAI14185.1 CM000160 EDW96511.1 GFDL01013356 JAV21689.1 GBYB01001889 JAG71656.1 IAAA01017739 LAA04751.1 AMQN01004769 AMQN01004770 KB294357 ELU14683.1 IAAA01017740 LAA04754.1 AAQR03172650 AJFE02046183 AACZ04055865 GABC01001118 GABF01004812 GABD01010132 GABE01009234 NBAG03000541 JAA10220.1 JAA17333.1 JAA22968.1 JAA35505.1 PNI16423.1 PNI16427.1 JRES01000196 KNC33363.1 ABLF02026893 CH902617 EDV43153.1 CH940650 EDW68065.1 GBYB01001892 JAG71659.1 GDIP01122697 JAL81017.1 GDIP01216625 JAJ06777.1 JXJN01008575 GDIP01120846 JAL82868.1 GAKP01008582 GAKP01008581 JAC50370.1 CP026245 AWO99309.1 JU323083 AFE66839.1 JSUE03022139 CH479179 EDW24378.1 JU472013 AFH28817.1
NWSH01000327 PCG77412.1 ODYU01006801 SOQ48990.1 AGBW02008899 OWR52233.1 PCG77413.1 JTDY01003451 KOB69536.1 GAIX01004168 JAA88392.1 KK852796 KDR16388.1 NEVH01003500 PNF40654.1 GL732533 EFX85136.1 GDIQ01257769 JAJ93955.1 GDIP01247258 LRGB01001579 JAI76143.1 KZS11549.1 KQ977121 KYN05544.1 KQ435713 KOX79322.1 KK107019 EZA62695.1 CH477195 EAT48401.1 KQ971354 EFA06186.1 AXCM01011152 GECZ01001504 JAS68265.1 ADMH02001047 ETN64329.1 GL452030 EFN78039.1 KQ981219 KYN44468.1 ATLV01027095 KE525423 KFB53895.1 AXCP01008618 AJVK01003341 ADTU01019796 KQ976450 KYM85732.1 GDRN01048555 JAI66647.1 GL768059 EFZ12716.1 GANO01001199 JAB58672.1 GEBQ01005948 JAT34029.1 KQ414727 KOC62530.1 PNF40655.1 AAZO01001857 DS235114 EEB12155.1 KZ288215 PBC32499.1 KQ435127 KZC14846.1 QCYY01002878 ROT66903.1 AJWK01001947 IACK01027967 LAA70675.1 KQ776210 OAD52211.1 UFQT01001048 SSX28617.1 NNAY01000508 OXU27943.1 GEZM01087528 JAV58645.1 KB202544 ESO89756.1 GBYB01001890 JAG71657.1 CP012526 ALC45460.1 GECZ01021734 JAS48035.1 UFQT01001411 SSX30433.1 GAMC01004649 JAC01907.1 FM956649 CAX11689.1 OUUW01000007 SPP82657.1 AXCN02001681 CH933806 EDW15508.1 DS232102 EDS34855.1 AKHW03002822 KYO37124.1 GBEX01000375 JAI14185.1 CM000160 EDW96511.1 GFDL01013356 JAV21689.1 GBYB01001889 JAG71656.1 IAAA01017739 LAA04751.1 AMQN01004769 AMQN01004770 KB294357 ELU14683.1 IAAA01017740 LAA04754.1 AAQR03172650 AJFE02046183 AACZ04055865 GABC01001118 GABF01004812 GABD01010132 GABE01009234 NBAG03000541 JAA10220.1 JAA17333.1 JAA22968.1 JAA35505.1 PNI16423.1 PNI16427.1 JRES01000196 KNC33363.1 ABLF02026893 CH902617 EDV43153.1 CH940650 EDW68065.1 GBYB01001892 JAG71659.1 GDIP01122697 JAL81017.1 GDIP01216625 JAJ06777.1 JXJN01008575 GDIP01120846 JAL82868.1 GAKP01008582 GAKP01008581 JAC50370.1 CP026245 AWO99309.1 JU323083 AFE66839.1 JSUE03022139 CH479179 EDW24378.1 JU472013 AFH28817.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000037510
+ More
UP000027135 UP000235965 UP000000305 UP000076858 UP000078542 UP000053105 UP000053097 UP000008820 UP000007266 UP000075884 UP000075883 UP000075920 UP000000673 UP000008237 UP000078541 UP000030765 UP000075880 UP000092462 UP000005205 UP000075881 UP000075900 UP000078540 UP000002358 UP000053825 UP000005203 UP000192223 UP000009046 UP000242457 UP000076502 UP000283509 UP000076408 UP000092461 UP000215335 UP000001646 UP000075885 UP000030746 UP000092553 UP000268350 UP000189705 UP000075886 UP000009192 UP000069272 UP000002320 UP000050525 UP000002282 UP000014760 UP000005225 UP000233180 UP000233200 UP000240080 UP000002277 UP000037069 UP000007819 UP000007801 UP000008792 UP000092460 UP000008225 UP000246464 UP000006718 UP000092445 UP000008744
UP000027135 UP000235965 UP000000305 UP000076858 UP000078542 UP000053105 UP000053097 UP000008820 UP000007266 UP000075884 UP000075883 UP000075920 UP000000673 UP000008237 UP000078541 UP000030765 UP000075880 UP000092462 UP000005205 UP000075881 UP000075900 UP000078540 UP000002358 UP000053825 UP000005203 UP000192223 UP000009046 UP000242457 UP000076502 UP000283509 UP000076408 UP000092461 UP000215335 UP000001646 UP000075885 UP000030746 UP000092553 UP000268350 UP000189705 UP000075886 UP000009192 UP000069272 UP000002320 UP000050525 UP000002282 UP000014760 UP000005225 UP000233180 UP000233200 UP000240080 UP000002277 UP000037069 UP000007819 UP000007801 UP000008792 UP000092460 UP000008225 UP000246464 UP000006718 UP000092445 UP000008744
PRIDE
Interpro
IPR027417
P-loop_NTPase
+ More
IPR013954 PNK3P
IPR041388 FHA_2
IPR006551 Polynucleotide_phosphatase
IPR008984 SMAD_FHA_dom_sf
IPR023214 HAD_sf
IPR006549 HAD-SF_hydro_IIIA
IPR036412 HAD-like_sf
IPR006550 PNKP
IPR000253 FHA_dom
IPR015047 DUF1866
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR002013 SAC_dom
IPR000300 IPPc
IPR036691 Endo/exonu/phosph_ase_sf
IPR035979 RBD_domain_sf
IPR034972 SYNJ1
IPR005135 Endo/exonuclease/phosphatase
IPR013954 PNK3P
IPR041388 FHA_2
IPR006551 Polynucleotide_phosphatase
IPR008984 SMAD_FHA_dom_sf
IPR023214 HAD_sf
IPR006549 HAD-SF_hydro_IIIA
IPR036412 HAD-like_sf
IPR006550 PNKP
IPR000253 FHA_dom
IPR015047 DUF1866
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR002013 SAC_dom
IPR000300 IPPc
IPR036691 Endo/exonu/phosph_ase_sf
IPR035979 RBD_domain_sf
IPR034972 SYNJ1
IPR005135 Endo/exonuclease/phosphatase
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9J782
A0A194QSY1
A0A194PTH4
A0A2A4K194
A0A2H1W7D6
A0A212FEU4
+ More
A0A2A4K0X3 A0A0L7L2G9 S4PXZ7 A0A067R0U4 A0A2J7RIJ5 E9G6T6 A0A0P5FHV9 A0A0P4X2H9 A0A151ILC0 A0A0N0BJP1 A0A026X339 Q17NZ7 D6WPR6 A0A182MXB4 A0A182MVD1 A0A182WDA0 A0A1B6H0S1 W5JNH4 E2C202 A0A195FVI1 A0A1S4EW42 A0A084WUK1 A0A182J3G3 A0A1B0D571 A0A158NLS9 A0A182JRT7 A0A182RAJ5 A0A195BLQ4 A0A0P4WQL4 E9J381 U5EV32 A0A1B6MDR6 K7IRQ0 A0A0L7QV67 A0A2J7RIJ1 A0A087ZW86 A0A1W4WYC1 E0VFJ9 A0A2A3EMX4 A0A154PSF1 A0A3R7LYR5 A0A182YQ91 A0A1B0C960 A0A2D4HFE4 A0A310SAT2 A0A336MJT2 A0A232FBV7 G1KMT2 A0A182PGC7 A0A1Y1KGB3 V4A8X4 A0A0C9R1P0 A0A0M4ENH1 A0A1B6FCW3 A0A336MNP9 W8C5W1 B8ZX67 A0A3B0JJY0 A0A1U7R7V0 A0A182QB07 B4K8T9 A0A182FTM2 B0WU84 A0A151NJZ6 A0A0F7ZAJ2 B4PT50 A0A1Q3F2C9 A0A0C9QEX3 A0A2L2Y9C8 R7V767 A0A2L2Y9D1 H0WRT3 A0A2K6JZR0 A0A2K6RGL1 A0A2R8ZCX7 K7BAU7 A0A0L0CLW8 J9JQT8 B3M0E7 B4LYW2 A0A0C9QMN2 A0A0P5V0A1 A0A0N7ZSL4 A0A1B0B4Z3 A0A0P5TWY5 A0A034W8B2 A0A2R8M3W8 A0A2U9B5U3 H9EWT0 F6PK17 A0A1A9ZT26 B4G3Q7 H9YYQ1
A0A2A4K0X3 A0A0L7L2G9 S4PXZ7 A0A067R0U4 A0A2J7RIJ5 E9G6T6 A0A0P5FHV9 A0A0P4X2H9 A0A151ILC0 A0A0N0BJP1 A0A026X339 Q17NZ7 D6WPR6 A0A182MXB4 A0A182MVD1 A0A182WDA0 A0A1B6H0S1 W5JNH4 E2C202 A0A195FVI1 A0A1S4EW42 A0A084WUK1 A0A182J3G3 A0A1B0D571 A0A158NLS9 A0A182JRT7 A0A182RAJ5 A0A195BLQ4 A0A0P4WQL4 E9J381 U5EV32 A0A1B6MDR6 K7IRQ0 A0A0L7QV67 A0A2J7RIJ1 A0A087ZW86 A0A1W4WYC1 E0VFJ9 A0A2A3EMX4 A0A154PSF1 A0A3R7LYR5 A0A182YQ91 A0A1B0C960 A0A2D4HFE4 A0A310SAT2 A0A336MJT2 A0A232FBV7 G1KMT2 A0A182PGC7 A0A1Y1KGB3 V4A8X4 A0A0C9R1P0 A0A0M4ENH1 A0A1B6FCW3 A0A336MNP9 W8C5W1 B8ZX67 A0A3B0JJY0 A0A1U7R7V0 A0A182QB07 B4K8T9 A0A182FTM2 B0WU84 A0A151NJZ6 A0A0F7ZAJ2 B4PT50 A0A1Q3F2C9 A0A0C9QEX3 A0A2L2Y9C8 R7V767 A0A2L2Y9D1 H0WRT3 A0A2K6JZR0 A0A2K6RGL1 A0A2R8ZCX7 K7BAU7 A0A0L0CLW8 J9JQT8 B3M0E7 B4LYW2 A0A0C9QMN2 A0A0P5V0A1 A0A0N7ZSL4 A0A1B0B4Z3 A0A0P5TWY5 A0A034W8B2 A0A2R8M3W8 A0A2U9B5U3 H9EWT0 F6PK17 A0A1A9ZT26 B4G3Q7 H9YYQ1
PDB
1YJ5
E-value=5.93548e-93,
Score=871
Ontologies
GO
GO:0016301
GO:0046403
GO:0006281
GO:0003690
GO:0046939
GO:0005634
GO:0046404
GO:0050145
GO:0098504
GO:0032212
GO:0005730
GO:0051973
GO:1904355
GO:0010836
GO:0006979
GO:0042769
GO:0042578
GO:0003676
GO:0046856
GO:0005515
GO:0006814
GO:0016020
GO:0008270
GO:0004812
GO:0006418
GO:0009058
GO:0016742
GO:0003824
PANTHER
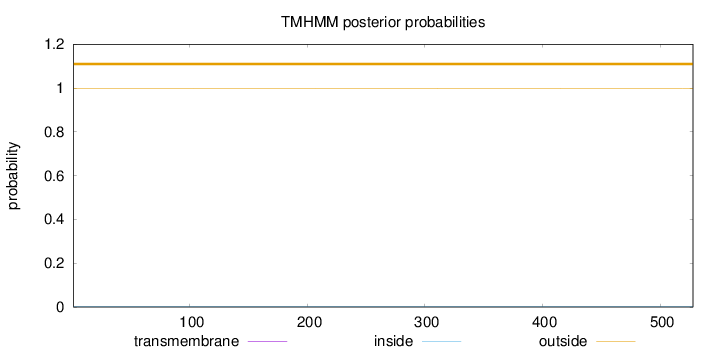
Topology
Length:
528
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0007
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00094
outside
1 - 528
Population Genetic Test Statistics
Pi
207.325654
Theta
192.799397
Tajima's D
0.628183
CLR
0.043573
CSRT
0.550722463876806
Interpretation
Uncertain
