Gene
KWMTBOMO04461
Pre Gene Modal
BGIBMGA005429
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_acylphosphatase-1-like_[Bombyx_mori]
Full name
Acylphosphatase
+ More
Acylphosphatase-1
Acylphosphatase-1
Alternative Name
Acylphosphate phosphohydrolase 1
Acylphosphatase, organ-common type isozyme
Acylphosphatase, organ-common type isozyme
Location in the cell
Cytoplasmic Reliability : 2.232
Sequence
CDS
ATGATGGCAGTGAACAAAGTAAATAGTGTAACGGCGGATTTCGAAGTTTTTGGAAAAGTGCAAGGAGTATTCTTCAGGAAGTTTACGCAAAAGAAAGCATTAGAACTTGGACTTAAGGGATGGGTAATGAATACGGCGCAAGGAACTGTTATAGGTCAACTTCAAGGACCCTCGACTGCTGTTGATGACATGAAGCTTTGGTTACAAAAGGTCGGAAGCCCAAAATCTAAGATCGAGAAAGCGGCGTTCAGGAATGAGGGAAATATTAAATGCTGCGCTTTTAGGACATTTGAAATACGTAAATGA
Protein
MMAVNKVNSVTADFEVFGKVQGVFFRKFTQKKALELGLKGWVMNTAQGTVIGQLQGPSTAVDDMKLWLQKVGSPKSKIEKAAFRNEGNIKCCAFRTFEIRK
Summary
Catalytic Activity
an acyl phosphate + H2O = a carboxylate + H(+) + phosphate
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Cofactor
Mg(2+)
Similarity
Belongs to the acylphosphatase family.
Keywords
Complete proteome
Hydrolase
Reference proteome
Acetylation
Alternative initiation
Direct protein sequencing
Feature
chain Acylphosphatase
splice variant In isoform ACYA.
splice variant In isoform ACYA.
Uniprot
A0A2H1WKS3
A0A0L7L3Z0
A0A194QSY6
A0A194PTH0
A0A154P670
E2C2U7
+ More
A0A194RME9 A0A194PM78 A0A151J0Y4 E1ZUY9 E9IIG8 A0A3L8DJ57 A0A151IBR8 A0A2A3ECT4 A0A088AM18 A0A151XHU8 T1IQP0 H3BG22 A0A212FLH6 A0A158NM38 A0A3B4B462 G1K3F8 B7ZSP5 A0A195B5B9 Q28FK7 A0A2H1V3B6 A0A232FK13 A0A151JV39 A0A0V0TJU1 A0A0V1D3B6 A0A0V1P7T1 A0A0V1JCU3 A0A0A9Y5B4 K7IUV6 A0A0V0XVY6 A0A0V1IXS4 A0A2P8YBJ7 A0A2G9RF53 B5G0Z0 A0A0L7RIB7 A0A3M7SEY4 A0A0C9R1Q2 A0A0V1D315 A0A0V0TJJ9 A0A0V1P7U8 A0A0V1D3G0 A0A0V0TJW4 A0A0V1P8G2 A0A0V0WKQ2 A0A0V0ZF83 A0A0V0SA42 A0A0V0SA05 A0A0V0S9Y9 A0A0V0S9Z5 A0A0V0SAZ7 A0A0V0SA69 A0A0V0SA20 B5G0Y8 A0A1S3J2T5 A0A2C9L6Q8 B5G0Y4 A0A060WLS8 B5G0Y6 A0A1L8F0L9 Q6DE05 A0A210PXJ7 A0A3Q2GRF1 W4Y3L6 B5G0Y7 A0A3Q3E0V9 M7BRE4 A0A2A4IZ47 A0A2A4IZ85 A0A1U8DBG4 H0ZPP1 A0A3Q2CJM0 A0A1V4KFQ5 A0A3Q3Q983 U3JV14 A0A1I8MYG9 G1NJC9 A0A3Q2ZE24 A0A3Q0RQ50 A0A0L0CG51 P24540 P24540-2 A0A085NMQ5 A0A1S3SPT5 A0A2P4SWE6 B3M373 A0A218VDH1 A0A077Z9X3 A0A0V1FSM1 A0A0V1MYP3 A0A0V1MY60 A0A1Z5LGQ1 A0A3S2M122 A0A2T7NYF4 B5G0Z3
A0A194RME9 A0A194PM78 A0A151J0Y4 E1ZUY9 E9IIG8 A0A3L8DJ57 A0A151IBR8 A0A2A3ECT4 A0A088AM18 A0A151XHU8 T1IQP0 H3BG22 A0A212FLH6 A0A158NM38 A0A3B4B462 G1K3F8 B7ZSP5 A0A195B5B9 Q28FK7 A0A2H1V3B6 A0A232FK13 A0A151JV39 A0A0V0TJU1 A0A0V1D3B6 A0A0V1P7T1 A0A0V1JCU3 A0A0A9Y5B4 K7IUV6 A0A0V0XVY6 A0A0V1IXS4 A0A2P8YBJ7 A0A2G9RF53 B5G0Z0 A0A0L7RIB7 A0A3M7SEY4 A0A0C9R1Q2 A0A0V1D315 A0A0V0TJJ9 A0A0V1P7U8 A0A0V1D3G0 A0A0V0TJW4 A0A0V1P8G2 A0A0V0WKQ2 A0A0V0ZF83 A0A0V0SA42 A0A0V0SA05 A0A0V0S9Y9 A0A0V0S9Z5 A0A0V0SAZ7 A0A0V0SA69 A0A0V0SA20 B5G0Y8 A0A1S3J2T5 A0A2C9L6Q8 B5G0Y4 A0A060WLS8 B5G0Y6 A0A1L8F0L9 Q6DE05 A0A210PXJ7 A0A3Q2GRF1 W4Y3L6 B5G0Y7 A0A3Q3E0V9 M7BRE4 A0A2A4IZ47 A0A2A4IZ85 A0A1U8DBG4 H0ZPP1 A0A3Q2CJM0 A0A1V4KFQ5 A0A3Q3Q983 U3JV14 A0A1I8MYG9 G1NJC9 A0A3Q2ZE24 A0A3Q0RQ50 A0A0L0CG51 P24540 P24540-2 A0A085NMQ5 A0A1S3SPT5 A0A2P4SWE6 B3M373 A0A218VDH1 A0A077Z9X3 A0A0V1FSM1 A0A0V1MYP3 A0A0V1MY60 A0A1Z5LGQ1 A0A3S2M122 A0A2T7NYF4 B5G0Z3
EC Number
3.6.1.7
Pubmed
EMBL
ODYU01009237
SOQ53512.1
JTDY01003170
KOB70019.1
KQ461154
KPJ08434.1
+ More
KQ459599 KPI94430.1 KQ434826 KZC07435.1 GL452203 EFN77782.1 KQ460205 KPJ17166.1 KQ459601 KPI94088.1 KQ980586 KYN15242.1 GL434335 EFN74987.1 GL763491 EFZ19644.1 QOIP01000007 RLU20381.1 KQ978079 KYM97165.1 KZ288282 PBC29565.1 KQ982109 KYQ59984.1 JH431312 AFYH01003232 AGBW02007799 OWR54550.1 ADTU01020221 AAMC01082889 BC170595 BC170599 AAI70595.1 KQ976604 KYM79384.1 CR761924 ODYU01000477 SOQ35333.1 NNAY01000115 OXU30839.1 KQ981730 KYN36606.1 JYDJ01000240 KRX39200.1 JYDI01000048 KRY55963.1 JYDM01000036 KRZ92266.1 JYDV01000108 KRZ32807.1 GBHO01016240 JAG27364.1 AAZX01010110 JYDU01000120 KRX92075.1 JYDS01000069 KRZ27543.1 PYGN01000725 PSN41638.1 KV943286 PIO26542.1 DQ215272 ACH44951.1 KQ414584 KOC70610.1 REGN01001486 RNA34384.1 GBYB01010064 JAG79831.1 KRY55964.1 KRX39199.1 KRZ92265.1 KRY55966.1 KRX39201.1 KRZ92267.1 JYDK01000102 KRX76369.1 JYDQ01000206 KRY11128.1 JYDL01000023 KRX23543.1 KRX23544.1 KRX23540.1 KRX23545.1 KRX23541.1 KRX23546.1 KRX23542.1 DQ215269 ACH44949.1 DQ215264 ACH44945.1 FR904607 CDQ68007.1 DQ215266 DQ215273 DQ215274 ACH44947.1 CM004481 OCT65132.1 BC077342 NEDP02005416 OWF41196.1 AAGJ04104659 DQ215268 DQ215270 ACH44948.1 KB548684 EMP30702.1 NWSH01005123 PCG64422.1 PCG64423.1 ABQF01003045 LSYS01003385 OPJ83235.1 AGTO01010448 JRES01000519 KNC30459.1 KL363220 KL367486 KFD53103.1 KFD70751.1 PPHD01019330 POI28437.1 CH902617 EDV43534.2 MUZQ01000005 OWK64115.1 HG806056 CDW56574.1 JYDT01000036 KRY89028.1 JYDO01000026 KRZ76707.1 KRZ76711.1 GFJQ02000387 JAW06583.1 RSAL01000081 RVE48579.1 PZQS01000008 PVD26184.1 DQ215275 ACH44954.1
KQ459599 KPI94430.1 KQ434826 KZC07435.1 GL452203 EFN77782.1 KQ460205 KPJ17166.1 KQ459601 KPI94088.1 KQ980586 KYN15242.1 GL434335 EFN74987.1 GL763491 EFZ19644.1 QOIP01000007 RLU20381.1 KQ978079 KYM97165.1 KZ288282 PBC29565.1 KQ982109 KYQ59984.1 JH431312 AFYH01003232 AGBW02007799 OWR54550.1 ADTU01020221 AAMC01082889 BC170595 BC170599 AAI70595.1 KQ976604 KYM79384.1 CR761924 ODYU01000477 SOQ35333.1 NNAY01000115 OXU30839.1 KQ981730 KYN36606.1 JYDJ01000240 KRX39200.1 JYDI01000048 KRY55963.1 JYDM01000036 KRZ92266.1 JYDV01000108 KRZ32807.1 GBHO01016240 JAG27364.1 AAZX01010110 JYDU01000120 KRX92075.1 JYDS01000069 KRZ27543.1 PYGN01000725 PSN41638.1 KV943286 PIO26542.1 DQ215272 ACH44951.1 KQ414584 KOC70610.1 REGN01001486 RNA34384.1 GBYB01010064 JAG79831.1 KRY55964.1 KRX39199.1 KRZ92265.1 KRY55966.1 KRX39201.1 KRZ92267.1 JYDK01000102 KRX76369.1 JYDQ01000206 KRY11128.1 JYDL01000023 KRX23543.1 KRX23544.1 KRX23540.1 KRX23545.1 KRX23541.1 KRX23546.1 KRX23542.1 DQ215269 ACH44949.1 DQ215264 ACH44945.1 FR904607 CDQ68007.1 DQ215266 DQ215273 DQ215274 ACH44947.1 CM004481 OCT65132.1 BC077342 NEDP02005416 OWF41196.1 AAGJ04104659 DQ215268 DQ215270 ACH44948.1 KB548684 EMP30702.1 NWSH01005123 PCG64422.1 PCG64423.1 ABQF01003045 LSYS01003385 OPJ83235.1 AGTO01010448 JRES01000519 KNC30459.1 KL363220 KL367486 KFD53103.1 KFD70751.1 PPHD01019330 POI28437.1 CH902617 EDV43534.2 MUZQ01000005 OWK64115.1 HG806056 CDW56574.1 JYDT01000036 KRY89028.1 JYDO01000026 KRZ76707.1 KRZ76711.1 GFJQ02000387 JAW06583.1 RSAL01000081 RVE48579.1 PZQS01000008 PVD26184.1 DQ215275 ACH44954.1
Proteomes
UP000037510
UP000053240
UP000053268
UP000076502
UP000008237
UP000078492
+ More
UP000000311 UP000279307 UP000078542 UP000242457 UP000005203 UP000075809 UP000008672 UP000007151 UP000005205 UP000261520 UP000008143 UP000078540 UP000215335 UP000078541 UP000055048 UP000054653 UP000054924 UP000054826 UP000002358 UP000054815 UP000054805 UP000245037 UP000053825 UP000276133 UP000054673 UP000054783 UP000054630 UP000085678 UP000076420 UP000193380 UP000186698 UP000242188 UP000265020 UP000007110 UP000261660 UP000031443 UP000218220 UP000189705 UP000007754 UP000190648 UP000261600 UP000016665 UP000095301 UP000001645 UP000264820 UP000261340 UP000037069 UP000008227 UP000087266 UP000007801 UP000197619 UP000030665 UP000054995 UP000054843 UP000283053 UP000245119
UP000000311 UP000279307 UP000078542 UP000242457 UP000005203 UP000075809 UP000008672 UP000007151 UP000005205 UP000261520 UP000008143 UP000078540 UP000215335 UP000078541 UP000055048 UP000054653 UP000054924 UP000054826 UP000002358 UP000054815 UP000054805 UP000245037 UP000053825 UP000276133 UP000054673 UP000054783 UP000054630 UP000085678 UP000076420 UP000193380 UP000186698 UP000242188 UP000265020 UP000007110 UP000261660 UP000031443 UP000218220 UP000189705 UP000007754 UP000190648 UP000261600 UP000016665 UP000095301 UP000001645 UP000264820 UP000261340 UP000037069 UP000008227 UP000087266 UP000007801 UP000197619 UP000030665 UP000054995 UP000054843 UP000283053 UP000245119
PRIDE
Interpro
IPR020456
Acylphosphatase
+ More
IPR001792 Acylphosphatase-like_dom
IPR036046 Acylphosphatase-like_dom_sf
IPR017968 Acylphosphatase_CS
IPR036412 HAD-like_sf
IPR023214 HAD_sf
IPR008380 HAD-SF_hydro_IG_5-nucl
IPR016695 Pur_nucleotidase
IPR003961 FN3_dom
IPR017441 Protein_kinase_ATP_BS
IPR013783 Ig-like_fold
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR036116 FN3_sf
IPR000719 Prot_kinase_dom
IPR020635 Tyr_kinase_cat_dom
IPR011044 Quino_amine_DH_bsu
IPR002011 Tyr_kinase_rcpt_2_CS
IPR011047 Quinoprotein_ADH-like_supfam
IPR008949 Isoprenoid_synthase_dom_sf
IPR001792 Acylphosphatase-like_dom
IPR036046 Acylphosphatase-like_dom_sf
IPR017968 Acylphosphatase_CS
IPR036412 HAD-like_sf
IPR023214 HAD_sf
IPR008380 HAD-SF_hydro_IG_5-nucl
IPR016695 Pur_nucleotidase
IPR003961 FN3_dom
IPR017441 Protein_kinase_ATP_BS
IPR013783 Ig-like_fold
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR036116 FN3_sf
IPR000719 Prot_kinase_dom
IPR020635 Tyr_kinase_cat_dom
IPR011044 Quino_amine_DH_bsu
IPR002011 Tyr_kinase_rcpt_2_CS
IPR011047 Quinoprotein_ADH-like_supfam
IPR008949 Isoprenoid_synthase_dom_sf
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A2H1WKS3
A0A0L7L3Z0
A0A194QSY6
A0A194PTH0
A0A154P670
E2C2U7
+ More
A0A194RME9 A0A194PM78 A0A151J0Y4 E1ZUY9 E9IIG8 A0A3L8DJ57 A0A151IBR8 A0A2A3ECT4 A0A088AM18 A0A151XHU8 T1IQP0 H3BG22 A0A212FLH6 A0A158NM38 A0A3B4B462 G1K3F8 B7ZSP5 A0A195B5B9 Q28FK7 A0A2H1V3B6 A0A232FK13 A0A151JV39 A0A0V0TJU1 A0A0V1D3B6 A0A0V1P7T1 A0A0V1JCU3 A0A0A9Y5B4 K7IUV6 A0A0V0XVY6 A0A0V1IXS4 A0A2P8YBJ7 A0A2G9RF53 B5G0Z0 A0A0L7RIB7 A0A3M7SEY4 A0A0C9R1Q2 A0A0V1D315 A0A0V0TJJ9 A0A0V1P7U8 A0A0V1D3G0 A0A0V0TJW4 A0A0V1P8G2 A0A0V0WKQ2 A0A0V0ZF83 A0A0V0SA42 A0A0V0SA05 A0A0V0S9Y9 A0A0V0S9Z5 A0A0V0SAZ7 A0A0V0SA69 A0A0V0SA20 B5G0Y8 A0A1S3J2T5 A0A2C9L6Q8 B5G0Y4 A0A060WLS8 B5G0Y6 A0A1L8F0L9 Q6DE05 A0A210PXJ7 A0A3Q2GRF1 W4Y3L6 B5G0Y7 A0A3Q3E0V9 M7BRE4 A0A2A4IZ47 A0A2A4IZ85 A0A1U8DBG4 H0ZPP1 A0A3Q2CJM0 A0A1V4KFQ5 A0A3Q3Q983 U3JV14 A0A1I8MYG9 G1NJC9 A0A3Q2ZE24 A0A3Q0RQ50 A0A0L0CG51 P24540 P24540-2 A0A085NMQ5 A0A1S3SPT5 A0A2P4SWE6 B3M373 A0A218VDH1 A0A077Z9X3 A0A0V1FSM1 A0A0V1MYP3 A0A0V1MY60 A0A1Z5LGQ1 A0A3S2M122 A0A2T7NYF4 B5G0Z3
A0A194RME9 A0A194PM78 A0A151J0Y4 E1ZUY9 E9IIG8 A0A3L8DJ57 A0A151IBR8 A0A2A3ECT4 A0A088AM18 A0A151XHU8 T1IQP0 H3BG22 A0A212FLH6 A0A158NM38 A0A3B4B462 G1K3F8 B7ZSP5 A0A195B5B9 Q28FK7 A0A2H1V3B6 A0A232FK13 A0A151JV39 A0A0V0TJU1 A0A0V1D3B6 A0A0V1P7T1 A0A0V1JCU3 A0A0A9Y5B4 K7IUV6 A0A0V0XVY6 A0A0V1IXS4 A0A2P8YBJ7 A0A2G9RF53 B5G0Z0 A0A0L7RIB7 A0A3M7SEY4 A0A0C9R1Q2 A0A0V1D315 A0A0V0TJJ9 A0A0V1P7U8 A0A0V1D3G0 A0A0V0TJW4 A0A0V1P8G2 A0A0V0WKQ2 A0A0V0ZF83 A0A0V0SA42 A0A0V0SA05 A0A0V0S9Y9 A0A0V0S9Z5 A0A0V0SAZ7 A0A0V0SA69 A0A0V0SA20 B5G0Y8 A0A1S3J2T5 A0A2C9L6Q8 B5G0Y4 A0A060WLS8 B5G0Y6 A0A1L8F0L9 Q6DE05 A0A210PXJ7 A0A3Q2GRF1 W4Y3L6 B5G0Y7 A0A3Q3E0V9 M7BRE4 A0A2A4IZ47 A0A2A4IZ85 A0A1U8DBG4 H0ZPP1 A0A3Q2CJM0 A0A1V4KFQ5 A0A3Q3Q983 U3JV14 A0A1I8MYG9 G1NJC9 A0A3Q2ZE24 A0A3Q0RQ50 A0A0L0CG51 P24540 P24540-2 A0A085NMQ5 A0A1S3SPT5 A0A2P4SWE6 B3M373 A0A218VDH1 A0A077Z9X3 A0A0V1FSM1 A0A0V1MYP3 A0A0V1MY60 A0A1Z5LGQ1 A0A3S2M122 A0A2T7NYF4 B5G0Z3
PDB
2ACY
E-value=7.01404e-23,
Score=258
Ontologies
PATHWAY
GO
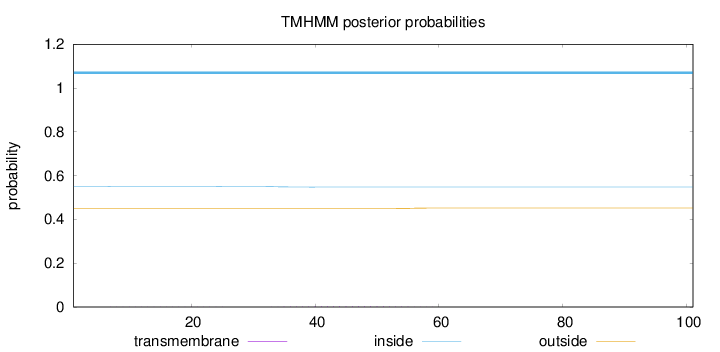
Topology
Length:
101
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06214
Exp number, first 60 AAs:
0.06208
Total prob of N-in:
0.55036
inside
1 - 101
Population Genetic Test Statistics
Pi
28.523609
Theta
31.288209
Tajima's D
-0.272402
CLR
2.490125
CSRT
0.301184940752962
Interpretation
Uncertain
