Gene
KWMTBOMO04459
Pre Gene Modal
BGIBMGA005376
Annotation
PREDICTED:_sodium_channel_protein_Nach-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.392 PlasmaMembrane Reliability : 1.78
Sequence
CDS
ATGACTAAGATCTTTTGGTTGGTATTCGTAGCACTCTCATGGTATGCTTCGTCATTACTGATAGCGGCTCAGTATGACGCGTATCTAAACAATCCTATTTCATTTGTGGTGGAAACAACTTACAAAGATTGGAATACATATTTTCCTGCCGTCGCTGTATGCGAAAACGACAATTCAAAACGTGCTGAAGAAATATCTGACAGTTTGTGGGGTCAAGAACATGATTTTAACATAGAGGAAGTTCTAAAGGAATGGGCATTTTTCAAAGGCGTCACATACTACACCTCGGAATTTTGTAATGTAGAAAATGGAGAAGTTTTACCGGAGTGCGTCGAATCTAATCTAACTTATTATGCCAATTTGGTAAGAAGTACCTGTAAAGAAAGTTTATCAGATTGTTCTTGGAACGGCGAAGCTTTTGAGTGTTGTGATTATTTCCGACCTTTGGATACTGAACTAGGCACGTGTTACGCCATCAACTCTATTCAAGGAAGAGAATCGAATCCGCCATTTCTACGAATGGTAAGCAACAGGACTACGGGTCCTGGAGTGCTGAGGTTCAATGTCCTAATACCCGCTAATGTTTACGTTCTAAACGAAGAAGACGTACCAGCTCTAACAACAATTGGATCAGATGTATTGACTGTACTGAAAGCACCATGGTCATCGAGGCCTGGTCTGTATTGCAACTGTTTACCGTCTTGTACAGAATCAGAGATTTCCGTCATTAATGATTTAAAAGACGCCACTGCCGAAGACTATGCCACTGTAGAAATAGAACTAGCGATGTTGCCATCCGAGCGGTACAGGAGGAATGTCGTAAGAGGAATACTTGACCTAGTCGGTTAG
Protein
MTKIFWLVFVALSWYASSLLIAAQYDAYLNNPISFVVETTYKDWNTYFPAVAVCENDNSKRAEEISDSLWGQEHDFNIEEVLKEWAFFKGVTYYTSEFCNVENGEVLPECVESNLTYYANLVRSTCKESLSDCSWNGEAFECCDYFRPLDTELGTCYAINSIQGRESNPPFLRMVSNRTTGPGVLRFNVLIPANVYVLNEEDVPALTTIGSDVLTVLKAPWSSRPGLYCNCLPSCTESEISVINDLKDATAEDYATVEIELAMLPSERYRRNVVRGILDLVG
Summary
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Uniprot
H9J784
A0A0L7L3I4
A0A194PNV6
A0A2H1WKA5
A0A194QTW1
B4MR50
+ More
B4GJH6 A0A3B0JMP3 B4P703 B3NKV3 B5DHT1 B4IFJ9 B4Q3Y0 Q86LG9 A0A1W4UV41 Q175M1 A0A182PJ85 A0A2M4CME2 A0A0M5J9K0 A0A0M5J227 A0A182YGS8 B3MU58 B4LUL8 W5JKJ9 A0A084VP06 A0A182KFM7 A0A1S4FE88 A0A182MU54 Q7Q3M0 A0A182WT66 B4JE99 A0A336MNG8 A0A336LWX4 A0A1I8N060 A0A1I8PKI8 A0A0L0C6A3 B0XIF0 U5ETC3 B4KKZ4 A0A182W9P5 A0A182RVK1 A0A194QXX8 D6WYQ4 A0A194PM81 A0A1W4WD96 A0A182NEB6 A0A1J1IPA5 A0A2A4K160 A0A2H1VYY4 A0A240SY07 A0A1B6DLH8 T1H8J9 J9K6V5 A0A1B6C2N5 A0A2J7RCQ7 A0A067RB98 A0A067QTA5 J9JYN8 A0A2J7PHM1 A0A182QV99 Q16L20 A0A1J1HFQ2 B4K758 A0A182GLK8 A0A1W4U653 A0A182GEY7 B0X7T4 A0A182UG85 A0A139WEM9 A0A084WH78 A0A1I8NTF7 Q176V2 A0A0L0BN58
B4GJH6 A0A3B0JMP3 B4P703 B3NKV3 B5DHT1 B4IFJ9 B4Q3Y0 Q86LG9 A0A1W4UV41 Q175M1 A0A182PJ85 A0A2M4CME2 A0A0M5J9K0 A0A0M5J227 A0A182YGS8 B3MU58 B4LUL8 W5JKJ9 A0A084VP06 A0A182KFM7 A0A1S4FE88 A0A182MU54 Q7Q3M0 A0A182WT66 B4JE99 A0A336MNG8 A0A336LWX4 A0A1I8N060 A0A1I8PKI8 A0A0L0C6A3 B0XIF0 U5ETC3 B4KKZ4 A0A182W9P5 A0A182RVK1 A0A194QXX8 D6WYQ4 A0A194PM81 A0A1W4WD96 A0A182NEB6 A0A1J1IPA5 A0A2A4K160 A0A2H1VYY4 A0A240SY07 A0A1B6DLH8 T1H8J9 J9K6V5 A0A1B6C2N5 A0A2J7RCQ7 A0A067RB98 A0A067QTA5 J9JYN8 A0A2J7PHM1 A0A182QV99 Q16L20 A0A1J1HFQ2 B4K758 A0A182GLK8 A0A1W4U653 A0A182GEY7 B0X7T4 A0A182UG85 A0A139WEM9 A0A084WH78 A0A1I8NTF7 Q176V2 A0A0L0BN58
Pubmed
EMBL
BABH01013660
JTDY01003170
KOB70022.1
KQ459599
KPI94429.1
ODYU01009237
+ More
SOQ53511.1 KQ461154 KPJ08435.1 CH963849 EDW74589.1 CH479184 EDW36792.1 OUUW01000006 SPP82133.1 CM000158 EDW89972.1 CH954179 EDV54476.1 CH379058 EDY69858.2 CH480833 EDW46421.1 CM000361 CM002910 EDX05696.1 KMY91271.1 AY226544 AE014134 AAO47370.1 AAX52675.1 CH477398 EAT41781.1 GGFL01002332 MBW66510.1 CP012523 ALC40356.1 ALC40344.1 CH902624 EDV33387.1 CH940649 EDW64204.1 ADMH02001256 ETN63400.1 ATLV01014965 KE524999 KFB39700.1 AXCM01001403 AAAB01008964 EAA12771.3 CH916368 EDW03619.1 UFQT01001837 SSX31962.1 UFQT01000264 SSX22556.1 JRES01000842 KNC27782.1 DS233296 EDS29204.1 GANO01002845 JAB57026.1 CH933807 EDW12744.1 KPJ08431.1 KQ971357 EFA08448.1 KPI94432.1 CVRI01000055 CRL01562.1 NWSH01000327 PCG77410.1 ODYU01005000 SOQ45444.1 AJWK01011557 GEDC01010828 JAS26470.1 ACPB03009340 ABLF02031349 ABLF02031358 GEDC01029779 JAS07519.1 NEVH01005884 PNF38621.1 KK852575 KDR21023.1 KK853042 KDR12140.1 ABLF02020828 NEVH01025135 PNF15837.1 AXCN02002523 CH477926 EAT35009.1 CVRI01000002 CRK86824.1 CH933806 EDW16371.1 JXUM01073172 KQ562760 KXJ75174.1 JXUM01058874 KQ562023 KXJ76884.1 DS232462 EDS42153.1 KQ971354 KYB26420.1 ATLV01023795 KE525346 KFB49572.1 CH477382 EAT42184.1 JRES01001623 KNC21456.1
SOQ53511.1 KQ461154 KPJ08435.1 CH963849 EDW74589.1 CH479184 EDW36792.1 OUUW01000006 SPP82133.1 CM000158 EDW89972.1 CH954179 EDV54476.1 CH379058 EDY69858.2 CH480833 EDW46421.1 CM000361 CM002910 EDX05696.1 KMY91271.1 AY226544 AE014134 AAO47370.1 AAX52675.1 CH477398 EAT41781.1 GGFL01002332 MBW66510.1 CP012523 ALC40356.1 ALC40344.1 CH902624 EDV33387.1 CH940649 EDW64204.1 ADMH02001256 ETN63400.1 ATLV01014965 KE524999 KFB39700.1 AXCM01001403 AAAB01008964 EAA12771.3 CH916368 EDW03619.1 UFQT01001837 SSX31962.1 UFQT01000264 SSX22556.1 JRES01000842 KNC27782.1 DS233296 EDS29204.1 GANO01002845 JAB57026.1 CH933807 EDW12744.1 KPJ08431.1 KQ971357 EFA08448.1 KPI94432.1 CVRI01000055 CRL01562.1 NWSH01000327 PCG77410.1 ODYU01005000 SOQ45444.1 AJWK01011557 GEDC01010828 JAS26470.1 ACPB03009340 ABLF02031349 ABLF02031358 GEDC01029779 JAS07519.1 NEVH01005884 PNF38621.1 KK852575 KDR21023.1 KK853042 KDR12140.1 ABLF02020828 NEVH01025135 PNF15837.1 AXCN02002523 CH477926 EAT35009.1 CVRI01000002 CRK86824.1 CH933806 EDW16371.1 JXUM01073172 KQ562760 KXJ75174.1 JXUM01058874 KQ562023 KXJ76884.1 DS232462 EDS42153.1 KQ971354 KYB26420.1 ATLV01023795 KE525346 KFB49572.1 CH477382 EAT42184.1 JRES01001623 KNC21456.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000053240
UP000007798
UP000008744
+ More
UP000268350 UP000002282 UP000008711 UP000001819 UP000001292 UP000000304 UP000000803 UP000192221 UP000008820 UP000075885 UP000092553 UP000076408 UP000007801 UP000008792 UP000000673 UP000030765 UP000075881 UP000075883 UP000007062 UP000076407 UP000001070 UP000095301 UP000095300 UP000037069 UP000002320 UP000009192 UP000075920 UP000075900 UP000007266 UP000192223 UP000075884 UP000183832 UP000218220 UP000092461 UP000015103 UP000007819 UP000235965 UP000027135 UP000075886 UP000069940 UP000249989 UP000075902
UP000268350 UP000002282 UP000008711 UP000001819 UP000001292 UP000000304 UP000000803 UP000192221 UP000008820 UP000075885 UP000092553 UP000076408 UP000007801 UP000008792 UP000000673 UP000030765 UP000075881 UP000075883 UP000007062 UP000076407 UP000001070 UP000095301 UP000095300 UP000037069 UP000002320 UP000009192 UP000075920 UP000075900 UP000007266 UP000192223 UP000075884 UP000183832 UP000218220 UP000092461 UP000015103 UP000007819 UP000235965 UP000027135 UP000075886 UP000069940 UP000249989 UP000075902
Interpro
IPR001873
ENaC
+ More
IPR015897 CHK_kinase-like
IPR004119 EcKinase-like
IPR011009 Kinase-like_dom_sf
IPR013087 Znf_C2H2_type
IPR036322 WD40_repeat_dom_sf
IPR036236 Znf_C2H2_sf
IPR012934 Znf_AD
IPR001680 WD40_repeat
IPR011047 Quinoprotein_ADH-like_supfam
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR009057 Homeobox-like_sf
IPR007889 HTH_Psq
IPR017896 4Fe4S_Fe-S-bd
IPR015897 CHK_kinase-like
IPR004119 EcKinase-like
IPR011009 Kinase-like_dom_sf
IPR013087 Znf_C2H2_type
IPR036322 WD40_repeat_dom_sf
IPR036236 Znf_C2H2_sf
IPR012934 Znf_AD
IPR001680 WD40_repeat
IPR011047 Quinoprotein_ADH-like_supfam
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR009057 Homeobox-like_sf
IPR007889 HTH_Psq
IPR017896 4Fe4S_Fe-S-bd
SUPFAM
Gene 3D
ProteinModelPortal
H9J784
A0A0L7L3I4
A0A194PNV6
A0A2H1WKA5
A0A194QTW1
B4MR50
+ More
B4GJH6 A0A3B0JMP3 B4P703 B3NKV3 B5DHT1 B4IFJ9 B4Q3Y0 Q86LG9 A0A1W4UV41 Q175M1 A0A182PJ85 A0A2M4CME2 A0A0M5J9K0 A0A0M5J227 A0A182YGS8 B3MU58 B4LUL8 W5JKJ9 A0A084VP06 A0A182KFM7 A0A1S4FE88 A0A182MU54 Q7Q3M0 A0A182WT66 B4JE99 A0A336MNG8 A0A336LWX4 A0A1I8N060 A0A1I8PKI8 A0A0L0C6A3 B0XIF0 U5ETC3 B4KKZ4 A0A182W9P5 A0A182RVK1 A0A194QXX8 D6WYQ4 A0A194PM81 A0A1W4WD96 A0A182NEB6 A0A1J1IPA5 A0A2A4K160 A0A2H1VYY4 A0A240SY07 A0A1B6DLH8 T1H8J9 J9K6V5 A0A1B6C2N5 A0A2J7RCQ7 A0A067RB98 A0A067QTA5 J9JYN8 A0A2J7PHM1 A0A182QV99 Q16L20 A0A1J1HFQ2 B4K758 A0A182GLK8 A0A1W4U653 A0A182GEY7 B0X7T4 A0A182UG85 A0A139WEM9 A0A084WH78 A0A1I8NTF7 Q176V2 A0A0L0BN58
B4GJH6 A0A3B0JMP3 B4P703 B3NKV3 B5DHT1 B4IFJ9 B4Q3Y0 Q86LG9 A0A1W4UV41 Q175M1 A0A182PJ85 A0A2M4CME2 A0A0M5J9K0 A0A0M5J227 A0A182YGS8 B3MU58 B4LUL8 W5JKJ9 A0A084VP06 A0A182KFM7 A0A1S4FE88 A0A182MU54 Q7Q3M0 A0A182WT66 B4JE99 A0A336MNG8 A0A336LWX4 A0A1I8N060 A0A1I8PKI8 A0A0L0C6A3 B0XIF0 U5ETC3 B4KKZ4 A0A182W9P5 A0A182RVK1 A0A194QXX8 D6WYQ4 A0A194PM81 A0A1W4WD96 A0A182NEB6 A0A1J1IPA5 A0A2A4K160 A0A2H1VYY4 A0A240SY07 A0A1B6DLH8 T1H8J9 J9K6V5 A0A1B6C2N5 A0A2J7RCQ7 A0A067RB98 A0A067QTA5 J9JYN8 A0A2J7PHM1 A0A182QV99 Q16L20 A0A1J1HFQ2 B4K758 A0A182GLK8 A0A1W4U653 A0A182GEY7 B0X7T4 A0A182UG85 A0A139WEM9 A0A084WH78 A0A1I8NTF7 Q176V2 A0A0L0BN58
Ontologies
GO
PANTHER
Topology
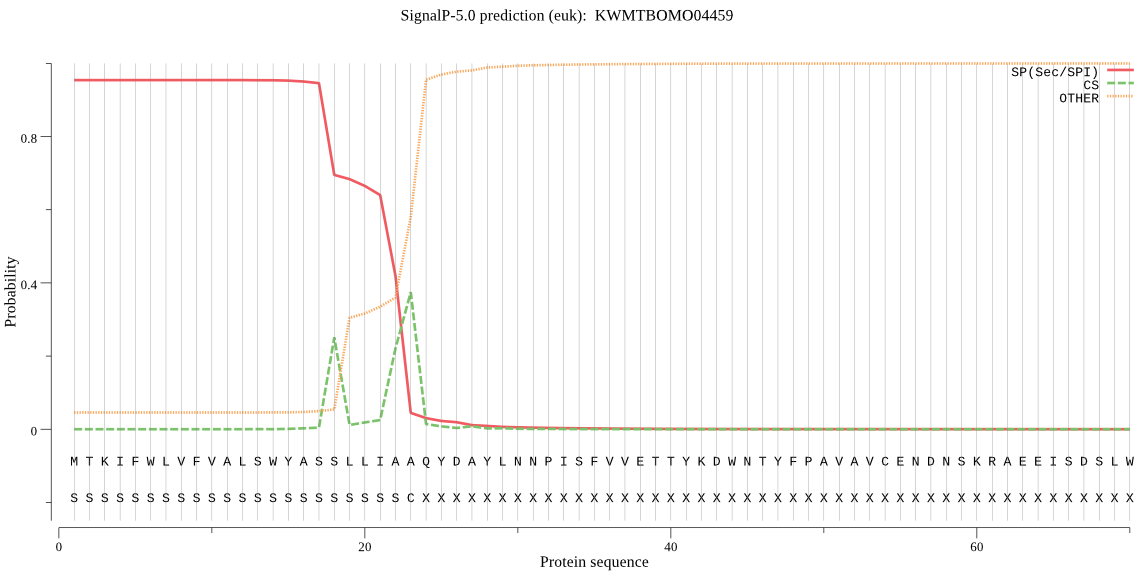
SignalP
Position: 1 - 23,
Likelihood: 0.953824
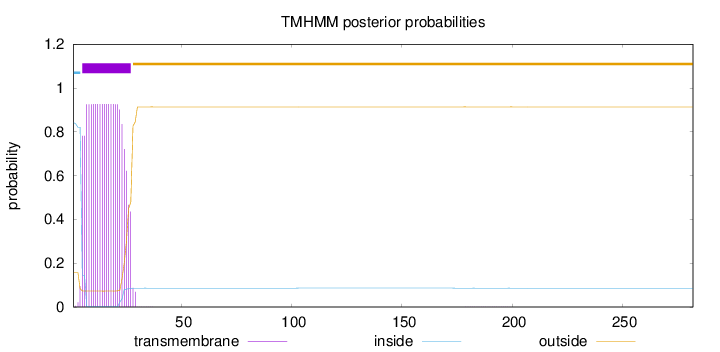
Length:
282
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.72914
Exp number, first 60 AAs:
19.7189
Total prob of N-in:
0.84208
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 282
Population Genetic Test Statistics
Pi
266.173863
Theta
185.469488
Tajima's D
1.089565
CLR
0.38893
CSRT
0.685815709214539
Interpretation
Uncertain
