Gene
KWMTBOMO04456
Pre Gene Modal
BGIBMGA005377
Annotation
PREDICTED:_general_transcription_factor_IIH_subunit_2_[Papilio_polytes]
Full name
General transcription factor IIH subunit
Location in the cell
Nuclear Reliability : 2.67
Sequence
CDS
ATGGCTGATGAAGATGACCCTAAAGAGTATCGATGGGAAACTGGTTATGAAAAAACTTGGGAGGCTATTAAAGAAGATGAAGATGGGTTGGTTGAAAGCCTGGTGGCTGAATTTGCCCAGAAAGCAGCACGTAAAGCAGCTGCTCCTCGTAAAGGTCCAGTCAGACTTGGCATGATGAGGCATTTACTTTTAGCCATAGATTGTTCTGAGGCTATGAGCTCCCAAGATTTGAAACCTTCACGGTTTCTGTGCACATTGAAGTTGCTCGAGAAATTTGTTGAAGAGTTTTTTGACCAGAATCCTCTCAGTCAGCTTGGTTTAATAGCAATGAAGAATAAGAGAGCTGAGAAAATAACTGACCTATCTGGAAATGTCCGGAAACACATTAAAGCCATACAAGGTTTATCCAACACTCCGCTGACTGGAGAGCCATCATTACAAAATACGCTAGAGCTGGCTGGGCGAATCCTAAGGCCTCTGCCTGGACACGCGTCAAGAGAATTACTAGTGCTTTTCGCATCCCTTACAACTTGCGATCCAAGTGATATAAACACTACCATAGAAACACTAAAAACAGATGGAATTAGATGTTCTGTTATTGGTCTAGCTGCTGAAGTGCGGATTTGCAAAAAAATGTGTCAAGACACTAATGGAGATTATGGTGTGGTATTAGACGATTGTCACTACCAGTCACTGCTACTAGAGTACACGTCGCCGCCGCCCCGCGCCCGCTCGTTGGACGCGGGCCTGGTGAAGATGGGCTTCCCGCATTCAACGCCGCTCGAGGAACAAGCCGACCCAGGGGCTAATGTCGATCCACCTATTACTGTTTGTATGTGTCATTTGGAGGAAGGCGAAGGTATAGGCGGCGAGGGTCACCTGTGTCCGCAGTGCCGCTCCAAGTATTGTTCGTTGCCAGCTCAGTGTCGCACCTGCGGCCTGACACTCGCATCAGCACCACATCTTGCAAGATCATACCATCACCTGTTTCCAGTGGAACCTTATGAAGAATTAGATAATGATGGCCAGGCGCAGTATTGTTTTGGATGCTTAAGAACCTTCACCGATATAGACAAGAAGATACATCGTTGTCCCCGATGTAAAGAGTACTACTGCTGGGAGTGCGAGGGCGCGGTGACGGCCACGCTGCACGTGTGTCCTGGCTGCGCCACCAGACCTCATTTGTACCAGCGGCTACCAAGCACGACCTCATCGTAA
Protein
MADEDDPKEYRWETGYEKTWEAIKEDEDGLVESLVAEFAQKAARKAAAPRKGPVRLGMMRHLLLAIDCSEAMSSQDLKPSRFLCTLKLLEKFVEEFFDQNPLSQLGLIAMKNKRAEKITDLSGNVRKHIKAIQGLSNTPLTGEPSLQNTLELAGRILRPLPGHASRELLVLFASLTTCDPSDINTTIETLKTDGIRCSVIGLAAEVRICKKMCQDTNGDYGVVLDDCHYQSLLLEYTSPPPRARSLDAGLVKMGFPHSTPLEEQADPGANVDPPITVCMCHLEEGEGIGGEGHLCPQCRSKYCSLPAQCRTCGLTLASAPHLARSYHHLFPVEPYEELDNDGQAQYCFGCLRTFTDIDKKIHRCPRCKEYYCWECEGAVTATLHVCPGCATRPHLYQRLPSTTSS
Summary
Similarity
Belongs to the GTF2H2 family.
Uniprot
A0A3S2NSH6
A0A2A4IYI8
A0A2H1VSI9
A0A194PMD0
H9J785
A0A194RD67
+ More
J3JTY9 A0A1Y1JS06 A0A336K9I8 B0WAT8 E2BY41 Q173R5 E2AW32 A0A026X2R0 A0A0L0CRY2 A0A2Z5TZB9 W8BSK5 A0A034WQM8 A0A1B0GNR2 A0A151WSC4 A0A1Q3F8S0 A0A1I8N6I9 A0A1I8QC55 A0A0A1XQC2 A0A1B0CJR7 A0A2J7R784 A0A151HY85 A0A0M9ABU6 W5JI03 A0A0K8VN72 A0A154NYT3 A0A0L7QXI3 A0A2J7R758 A0A182FUK7 A0A067RH91 A0A182KCD8 A0A0M4EH85 A0A2A3ET46 A0A182QQ13 A0A088A575 A0A151I792 K7IVF9 A0A182X1V8 A0A182V099 A0A182LQ16 Q7PPS5 A0A182HFS2 F4X4X4 A0A182P8I0 A0A182MYD0 A0A182IQB4 A0A182UAR1 A0A182VTB1 A0A1A9XBJ9 A0A182MLR7 A0A310SHB4 A0A182YSS7 B4KYL1 A0A182RLQ3 B4J3S1 B4LDP4 A0A0A9YGM3 B4GRB7 Q2M0P7 A0A0J7KN82 A0A3B0K247 T1JJK0 B3MBA0 A0A1B6DUF4 A0A1J1HS22 B4N3K4 A0A084VKX6 A0A1A9WS76 A0A0P5W7H0 E9GFE0 B4PIR4 A0A0N8E1E5 A0A1W4UE88 B4QLB8 Q9VNP8 A0A0P5B7H2 A0A210PXN6 V4B2E9 B4IB56 B3NEG9 A0A1A9UXV1 A0A1B0BAD5 A0A1A9Z3L9 D3TRH4 K1RAW7 A0A158NVZ4 A0A023F313 A0A0P5VYR5 A0A0P6I4D1 A0A0P4ZW49 A0A2P6LCB3 E0VFY0 A0A087TNQ8
J3JTY9 A0A1Y1JS06 A0A336K9I8 B0WAT8 E2BY41 Q173R5 E2AW32 A0A026X2R0 A0A0L0CRY2 A0A2Z5TZB9 W8BSK5 A0A034WQM8 A0A1B0GNR2 A0A151WSC4 A0A1Q3F8S0 A0A1I8N6I9 A0A1I8QC55 A0A0A1XQC2 A0A1B0CJR7 A0A2J7R784 A0A151HY85 A0A0M9ABU6 W5JI03 A0A0K8VN72 A0A154NYT3 A0A0L7QXI3 A0A2J7R758 A0A182FUK7 A0A067RH91 A0A182KCD8 A0A0M4EH85 A0A2A3ET46 A0A182QQ13 A0A088A575 A0A151I792 K7IVF9 A0A182X1V8 A0A182V099 A0A182LQ16 Q7PPS5 A0A182HFS2 F4X4X4 A0A182P8I0 A0A182MYD0 A0A182IQB4 A0A182UAR1 A0A182VTB1 A0A1A9XBJ9 A0A182MLR7 A0A310SHB4 A0A182YSS7 B4KYL1 A0A182RLQ3 B4J3S1 B4LDP4 A0A0A9YGM3 B4GRB7 Q2M0P7 A0A0J7KN82 A0A3B0K247 T1JJK0 B3MBA0 A0A1B6DUF4 A0A1J1HS22 B4N3K4 A0A084VKX6 A0A1A9WS76 A0A0P5W7H0 E9GFE0 B4PIR4 A0A0N8E1E5 A0A1W4UE88 B4QLB8 Q9VNP8 A0A0P5B7H2 A0A210PXN6 V4B2E9 B4IB56 B3NEG9 A0A1A9UXV1 A0A1B0BAD5 A0A1A9Z3L9 D3TRH4 K1RAW7 A0A158NVZ4 A0A023F313 A0A0P5VYR5 A0A0P6I4D1 A0A0P4ZW49 A0A2P6LCB3 E0VFY0 A0A087TNQ8
Pubmed
26354079
19121390
22516182
23537049
28004739
20798317
+ More
17510324 24508170 30249741 26108605 26760975 24495485 25348373 25315136 25830018 20920257 23761445 24845553 20075255 20966253 12364791 14747013 17210077 21719571 25244985 17994087 25401762 15632085 23185243 24438588 21292972 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28812685 23254933 20353571 22992520 21347285 25474469 20566863
17510324 24508170 30249741 26108605 26760975 24495485 25348373 25315136 25830018 20920257 23761445 24845553 20075255 20966253 12364791 14747013 17210077 21719571 25244985 17994087 25401762 15632085 23185243 24438588 21292972 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28812685 23254933 20353571 22992520 21347285 25474469 20566863
EMBL
RSAL01000258
RVE43372.1
NWSH01005050
PCG64478.1
ODYU01003876
SOQ43184.1
+ More
KQ459599 KPI94477.1 BABH01013657 KQ460397 KPJ15220.1 APGK01057992 APGK01057993 APGK01057994 APGK01057995 BT126699 KB741285 KB632425 AEE61661.1 ENN70742.1 ERL95353.1 GEZM01102799 JAV51783.1 UFQS01000195 UFQT01000195 SSX01097.1 SSX21477.1 DS231875 EDS41804.1 GL451420 EFN79320.1 CH477418 EAT41272.1 GL443213 EFN62454.1 KK107020 QOIP01000006 EZA62580.1 RLU21850.1 JRES01000005 KNC34981.1 FX985775 BBA93662.1 GAMC01004438 JAC02118.1 GAKP01002457 JAC56495.1 AJVK01013549 KQ982782 KYQ50728.1 GFDL01011157 JAV23888.1 GBXI01001559 JAD12733.1 AJWK01014895 NEVH01006738 PNF36666.1 KQ976730 KYM76404.1 KQ435687 KOX81345.1 ADMH02001479 ETN62399.1 GDHF01011988 JAI40326.1 KQ434783 KZC04783.1 KQ414705 KOC63249.1 PNF36669.1 KK852470 KDR23206.1 CP012525 ALC43038.1 KZ288191 PBC34472.1 AXCN02002052 KQ978426 KYM94032.1 AAAB01008933 EAA09916.5 APCN01005568 GL888679 EGI58547.1 AXCM01003009 KQ759893 OAD62067.1 CH933809 EDW18822.1 CH916366 EDV97302.1 CH940647 EDW70005.1 GBHO01014964 GBRD01001178 JAG28640.1 JAG64643.1 CH479188 EDW40302.1 CH379069 EAL30883.2 LBMM01005164 KMQ91772.1 OUUW01000012 SPP87393.1 JH432085 CH902618 EDV41401.1 GEDC01008029 JAS29269.1 CVRI01000011 CRK89278.1 CH964095 EDW79209.2 ATLV01014310 KE524956 KFB38620.1 GDIP01090354 JAM13361.1 GL732542 EFX81828.1 CM000159 EDW95572.1 GDIQ01071308 JAN23429.1 CM000363 CM002912 EDX11536.1 KMZ01251.1 AE014296 AY047574 AAF51879.2 AAK77306.1 GDIP01188387 JAJ35015.1 NEDP02005415 OWF41247.1 KB203855 ESO82529.1 CH480826 EDW44519.1 CH954178 EDV52804.1 JXJN01010916 CCAG010018371 EZ424026 ADD20302.1 JH818113 EKC40804.1 ADTU01027567 GBBI01003278 JAC15434.1 GDIP01093299 LRGB01000389 JAM10416.1 KZS19422.1 GDIQ01009688 JAN85049.1 GDIP01207939 JAJ15463.1 MWRG01000400 PRD36201.1 DS235129 EEB12286.1 KK116084 KFM66747.1
KQ459599 KPI94477.1 BABH01013657 KQ460397 KPJ15220.1 APGK01057992 APGK01057993 APGK01057994 APGK01057995 BT126699 KB741285 KB632425 AEE61661.1 ENN70742.1 ERL95353.1 GEZM01102799 JAV51783.1 UFQS01000195 UFQT01000195 SSX01097.1 SSX21477.1 DS231875 EDS41804.1 GL451420 EFN79320.1 CH477418 EAT41272.1 GL443213 EFN62454.1 KK107020 QOIP01000006 EZA62580.1 RLU21850.1 JRES01000005 KNC34981.1 FX985775 BBA93662.1 GAMC01004438 JAC02118.1 GAKP01002457 JAC56495.1 AJVK01013549 KQ982782 KYQ50728.1 GFDL01011157 JAV23888.1 GBXI01001559 JAD12733.1 AJWK01014895 NEVH01006738 PNF36666.1 KQ976730 KYM76404.1 KQ435687 KOX81345.1 ADMH02001479 ETN62399.1 GDHF01011988 JAI40326.1 KQ434783 KZC04783.1 KQ414705 KOC63249.1 PNF36669.1 KK852470 KDR23206.1 CP012525 ALC43038.1 KZ288191 PBC34472.1 AXCN02002052 KQ978426 KYM94032.1 AAAB01008933 EAA09916.5 APCN01005568 GL888679 EGI58547.1 AXCM01003009 KQ759893 OAD62067.1 CH933809 EDW18822.1 CH916366 EDV97302.1 CH940647 EDW70005.1 GBHO01014964 GBRD01001178 JAG28640.1 JAG64643.1 CH479188 EDW40302.1 CH379069 EAL30883.2 LBMM01005164 KMQ91772.1 OUUW01000012 SPP87393.1 JH432085 CH902618 EDV41401.1 GEDC01008029 JAS29269.1 CVRI01000011 CRK89278.1 CH964095 EDW79209.2 ATLV01014310 KE524956 KFB38620.1 GDIP01090354 JAM13361.1 GL732542 EFX81828.1 CM000159 EDW95572.1 GDIQ01071308 JAN23429.1 CM000363 CM002912 EDX11536.1 KMZ01251.1 AE014296 AY047574 AAF51879.2 AAK77306.1 GDIP01188387 JAJ35015.1 NEDP02005415 OWF41247.1 KB203855 ESO82529.1 CH480826 EDW44519.1 CH954178 EDV52804.1 JXJN01010916 CCAG010018371 EZ424026 ADD20302.1 JH818113 EKC40804.1 ADTU01027567 GBBI01003278 JAC15434.1 GDIP01093299 LRGB01000389 JAM10416.1 KZS19422.1 GDIQ01009688 JAN85049.1 GDIP01207939 JAJ15463.1 MWRG01000400 PRD36201.1 DS235129 EEB12286.1 KK116084 KFM66747.1
Proteomes
UP000283053
UP000218220
UP000053268
UP000005204
UP000053240
UP000019118
+ More
UP000030742 UP000002320 UP000008237 UP000008820 UP000000311 UP000053097 UP000279307 UP000037069 UP000092462 UP000075809 UP000095301 UP000095300 UP000092461 UP000235965 UP000078540 UP000053105 UP000000673 UP000076502 UP000053825 UP000069272 UP000027135 UP000075881 UP000092553 UP000242457 UP000075886 UP000005203 UP000078542 UP000002358 UP000076407 UP000075903 UP000075882 UP000007062 UP000075840 UP000007755 UP000075885 UP000075884 UP000075880 UP000075902 UP000075920 UP000092443 UP000075883 UP000076408 UP000009192 UP000075900 UP000001070 UP000008792 UP000008744 UP000001819 UP000036403 UP000268350 UP000007801 UP000183832 UP000007798 UP000030765 UP000091820 UP000000305 UP000002282 UP000192221 UP000000304 UP000000803 UP000242188 UP000030746 UP000001292 UP000008711 UP000078200 UP000092460 UP000092445 UP000092444 UP000005408 UP000005205 UP000076858 UP000009046 UP000054359
UP000030742 UP000002320 UP000008237 UP000008820 UP000000311 UP000053097 UP000279307 UP000037069 UP000092462 UP000075809 UP000095301 UP000095300 UP000092461 UP000235965 UP000078540 UP000053105 UP000000673 UP000076502 UP000053825 UP000069272 UP000027135 UP000075881 UP000092553 UP000242457 UP000075886 UP000005203 UP000078542 UP000002358 UP000076407 UP000075903 UP000075882 UP000007062 UP000075840 UP000007755 UP000075885 UP000075884 UP000075880 UP000075902 UP000075920 UP000092443 UP000075883 UP000076408 UP000009192 UP000075900 UP000001070 UP000008792 UP000008744 UP000001819 UP000036403 UP000268350 UP000007801 UP000183832 UP000007798 UP000030765 UP000091820 UP000000305 UP000002282 UP000192221 UP000000304 UP000000803 UP000242188 UP000030746 UP000001292 UP000008711 UP000078200 UP000092460 UP000092445 UP000092444 UP000005408 UP000005205 UP000076858 UP000009046 UP000054359
Interpro
SUPFAM
SSF53300
SSF53300
Gene 3D
ProteinModelPortal
A0A3S2NSH6
A0A2A4IYI8
A0A2H1VSI9
A0A194PMD0
H9J785
A0A194RD67
+ More
J3JTY9 A0A1Y1JS06 A0A336K9I8 B0WAT8 E2BY41 Q173R5 E2AW32 A0A026X2R0 A0A0L0CRY2 A0A2Z5TZB9 W8BSK5 A0A034WQM8 A0A1B0GNR2 A0A151WSC4 A0A1Q3F8S0 A0A1I8N6I9 A0A1I8QC55 A0A0A1XQC2 A0A1B0CJR7 A0A2J7R784 A0A151HY85 A0A0M9ABU6 W5JI03 A0A0K8VN72 A0A154NYT3 A0A0L7QXI3 A0A2J7R758 A0A182FUK7 A0A067RH91 A0A182KCD8 A0A0M4EH85 A0A2A3ET46 A0A182QQ13 A0A088A575 A0A151I792 K7IVF9 A0A182X1V8 A0A182V099 A0A182LQ16 Q7PPS5 A0A182HFS2 F4X4X4 A0A182P8I0 A0A182MYD0 A0A182IQB4 A0A182UAR1 A0A182VTB1 A0A1A9XBJ9 A0A182MLR7 A0A310SHB4 A0A182YSS7 B4KYL1 A0A182RLQ3 B4J3S1 B4LDP4 A0A0A9YGM3 B4GRB7 Q2M0P7 A0A0J7KN82 A0A3B0K247 T1JJK0 B3MBA0 A0A1B6DUF4 A0A1J1HS22 B4N3K4 A0A084VKX6 A0A1A9WS76 A0A0P5W7H0 E9GFE0 B4PIR4 A0A0N8E1E5 A0A1W4UE88 B4QLB8 Q9VNP8 A0A0P5B7H2 A0A210PXN6 V4B2E9 B4IB56 B3NEG9 A0A1A9UXV1 A0A1B0BAD5 A0A1A9Z3L9 D3TRH4 K1RAW7 A0A158NVZ4 A0A023F313 A0A0P5VYR5 A0A0P6I4D1 A0A0P4ZW49 A0A2P6LCB3 E0VFY0 A0A087TNQ8
J3JTY9 A0A1Y1JS06 A0A336K9I8 B0WAT8 E2BY41 Q173R5 E2AW32 A0A026X2R0 A0A0L0CRY2 A0A2Z5TZB9 W8BSK5 A0A034WQM8 A0A1B0GNR2 A0A151WSC4 A0A1Q3F8S0 A0A1I8N6I9 A0A1I8QC55 A0A0A1XQC2 A0A1B0CJR7 A0A2J7R784 A0A151HY85 A0A0M9ABU6 W5JI03 A0A0K8VN72 A0A154NYT3 A0A0L7QXI3 A0A2J7R758 A0A182FUK7 A0A067RH91 A0A182KCD8 A0A0M4EH85 A0A2A3ET46 A0A182QQ13 A0A088A575 A0A151I792 K7IVF9 A0A182X1V8 A0A182V099 A0A182LQ16 Q7PPS5 A0A182HFS2 F4X4X4 A0A182P8I0 A0A182MYD0 A0A182IQB4 A0A182UAR1 A0A182VTB1 A0A1A9XBJ9 A0A182MLR7 A0A310SHB4 A0A182YSS7 B4KYL1 A0A182RLQ3 B4J3S1 B4LDP4 A0A0A9YGM3 B4GRB7 Q2M0P7 A0A0J7KN82 A0A3B0K247 T1JJK0 B3MBA0 A0A1B6DUF4 A0A1J1HS22 B4N3K4 A0A084VKX6 A0A1A9WS76 A0A0P5W7H0 E9GFE0 B4PIR4 A0A0N8E1E5 A0A1W4UE88 B4QLB8 Q9VNP8 A0A0P5B7H2 A0A210PXN6 V4B2E9 B4IB56 B3NEG9 A0A1A9UXV1 A0A1B0BAD5 A0A1A9Z3L9 D3TRH4 K1RAW7 A0A158NVZ4 A0A023F313 A0A0P5VYR5 A0A0P6I4D1 A0A0P4ZW49 A0A2P6LCB3 E0VFY0 A0A087TNQ8
PDB
5O85
E-value=7.76501e-99,
Score=920
Ontologies
PATHWAY
GO
Topology
Subcellular location
Nucleus
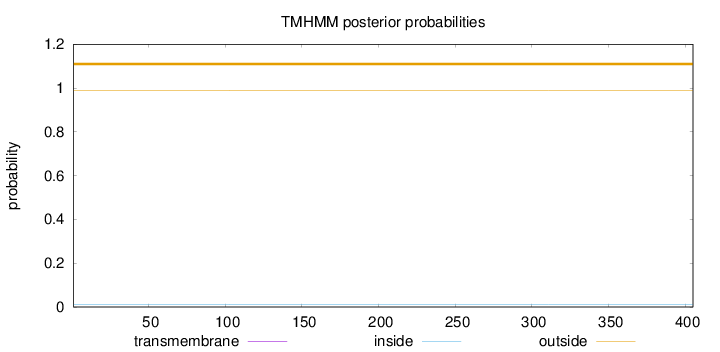
Length:
405
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.001
Exp number, first 60 AAs:
0.00021
Total prob of N-in:
0.00979
outside
1 - 405
Population Genetic Test Statistics
Pi
239.257879
Theta
174.418219
Tajima's D
0.979748
CLR
0.149381
CSRT
0.655967201639918
Interpretation
Uncertain
