Gene
KWMTBOMO04455
Pre Gene Modal
BGIBMGA005424
Annotation
hypothetical_protein_KGM_06855_[Danaus_plexippus]
Full name
Protein kinase C
Location in the cell
PlasmaMembrane Reliability : 4.94
Sequence
CDS
ATGAAATGCGATGGAGCCCACGAAGCGATGCTCGGCAATGACAAATTGGCGAAAATCGCTAAACAAAAGCAGAAAGAATCAAATAAACAGGACGAATACGAAGCCCTACAGAGTTTCCTGCGCAGTATCAGCTCAACGGCCACATTACCACCATACGTCAAAGGAGAAACATACTCTTCCGTTCGGGGTGTATTCAATCAGTGCCTCATAACATGTGCAGTGTTGATCCTTGCCGCCGGTGCGGGTCATCCTATCGGTTTCTCAGCGATTGCTCTACCACAACTGCAGCGTGAGAATAGCTCTATGAGAACAGACGACGAGATGGGCTCTTGGATCGCCAGTATCCACTCGGCAGCTACCCCTCTCGGCTCGATGGTGTCAGGTCCGGTGATGGAAGCCATCGGGCGGCGGCGGACGCTACAGTCCTGCACCGTCCCGCTGGTCGTTGGGTGGATCATAATAGGAACTGCGTCGCATCACGCTTTATTGCTCCTTGGTAGAATTGTCTGTGGCTTCGCTGTAGGCCTCATGGCCGCTCCGTCACAGGTCTATTTGGGTGAAATATCTGAGCCCAGATTACGAGGTCTACTAATTGGAACTCCGTTTGTTGCATATTCTCTTGGAGTGTTGTACGTTTACACCCTGGGCGGTCCCCTTAGTTGGAGGTCCGTTGCATATCTCTCCACTGTACTCCCAATATTAGCATTTATTGCTCTATGCTTCTCTCCGGAAAGCCCGACATGGTTGGCTAGAAGAGGAAGATTCCACGAGGCCATGGCAGCGATGTCACGACTCCGAGGCACTACTGAAACGGCACAACGAGAACTCCATGAATTAATATCATGTCGGGAGAAGGAGAAAGCGCGAGGGGAAGAGAATATCAAATTCTTCGCAACTGTCATTCGATCCCCTGTGTTGAAACCACTGATTCTGATCAACACGTTCAACTTACTGCAGATTCTGTCAGGAAGTTATGTCGTTATATTTTATGCCATCAAAATTGTTGAAGAAGCCGGCGGTTCTCTGCCGCCTGAGATGGCAGCCAACGCAAGCGCTTTAGTAAGGTTAGCAGTGACGATCCTCGCTTGCATCTTGTTACTGCGGCTCACTCGTCGGACACTCGTCATGATTTCTGGCATTGGAACTTCTGTCTGTACTCTGGCGTTGGCCGCCCTACTCGCGCAAGGCCCCGGTACAGGCATCGCGCCGCCAGTCCTGATCCTCGCATACGTTGGCTTCAATACCTTAGGATTCTTTGTACTGCCGGGGCTTATGATAGGCGAATTACTGCCCACCAAAGTCCGTGGGCTCTGCGGAGGCTATATTTTCTGTTTCTTTAATTCATTCCTTTTTGGTTTTACCAAACTTTACCCTGCGATGATGAAGAATATAGGAATAGCGGCTGTCTTTAGTTTCTTCGGTATTTCCGCTGCTCTTGCCACAATTCTACTTTTCCTTTTATTACCAGAGACGAAAGGGAAATCTCTTTTACAAATAGAACAGTATTATCAAAAGCCGAATATATTGTGGCTGTCTCGTAAAAAAACAACCGGTAGTCAAAACGTTTGA
Protein
MKCDGAHEAMLGNDKLAKIAKQKQKESNKQDEYEALQSFLRSISSTATLPPYVKGETYSSVRGVFNQCLITCAVLILAAGAGHPIGFSAIALPQLQRENSSMRTDDEMGSWIASIHSAATPLGSMVSGPVMEAIGRRRTLQSCTVPLVVGWIIIGTASHHALLLLGRIVCGFAVGLMAAPSQVYLGEISEPRLRGLLIGTPFVAYSLGVLYVYTLGGPLSWRSVAYLSTVLPILAFIALCFSPESPTWLARRGRFHEAMAAMSRLRGTTETAQRELHELISCREKEKARGEENIKFFATVIRSPVLKPLILINTFNLLQILSGSYVVIFYAIKIVEEAGGSLPPEMAANASALVRLAVTILACILLLRLTRRTLVMISGIGTSVCTLALAALLAQGPGTGIAPPVLILAYVGFNTLGFFVLPGLMIGELLPTKVRGLCGGYIFCFFNSFLFGFTKLYPAMMKNIGIAAVFSFFGISAALATILLFLLLPETKGKSLLQIEQYYQKPNILWLSRKKTTGSQNV
Summary
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Similarity
Belongs to the membrane-bound acyltransferase family.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. PKC subfamily.
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. PKC subfamily.
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9J7D2
A0A212F561
A0A3S2NBT7
A0A2H1VQR6
A0A194RH13
A0A194PNB3
+ More
A0A2A4IZE6 A0A182JVB0 A0A182IM61 A0A182RGI9 A0A182QYL2 A0A1S4FT49 A0A182Y1Z9 A0A182PRA4 A0A1B0CEP4 A0A1L8DFD0 A0A182M1R2 A0A1L8DF83 E2A4J1 A0A182WZA8 A0A1C7CZM0 A0A182NPD2 T1PIT2 A0A1I8NGC6 A0A182HPW0 A0A182U1D7 A0A182UQN7 A0A026W393 A0A182UBV2 A0A182FW58 A0A182SB54 W5JJV0 Q7Q024 A0A2J7QE56 A0A2M4AEH4 D6X016 A0A2M4AEJ1 A0A2M4AEY6 A0A2P8XF39 A0A2M4CN26 A0A0L0CQE9 A0A1I8NN12 A0A2M4AFX6 A0A1A9XP26 A0A1B0BNI6 E2B9R1 A0A151WLJ9 A0A1B0G7Q1 A0A1A9ZWD8 A0A1A9V8D1 A0A2M4BIF5 A0A182W9C7 A0A151IXJ2 A0A2M4BLV3 A0A195FQL9 B4PA70 B4QAZ8 A0A1I8M1P2 Q8MLQ7 W8CAE6 B3NPS7 A0A195CU57 A0A1Y1L0W9 A0A1Q3G0C6 B4I8T7 Q0IEA9 A0A0A1XJU0 F4WIX4 A0A034WQX0 A0A084WHI6 B4GBK8 A0A158N976 A0A3B0JXX9 B4LIZ5 A0A1J1ICI9 A0A0K8VSF9 B4KUC7 A0A1W4UMN4 A0A1W4V1P0 B4MYD6 B3ME82 A0A1Y1KX95 A0A1A9WIQ7 A0A0M4EV96 A0A0J7L656 B4JVU0 A0A0M9ADJ5 Q9W1J2 A0A0L7QJV6 A0A195B4K6 A0A2J7QE64 A0A182GZS3 A0A3L8DA70 A0A0A8J8P2 A0A1I8NN11 A0A067RK37 A0A1L8DDZ9 A0A1L8DDX2 A0A1Y1JT67
A0A2A4IZE6 A0A182JVB0 A0A182IM61 A0A182RGI9 A0A182QYL2 A0A1S4FT49 A0A182Y1Z9 A0A182PRA4 A0A1B0CEP4 A0A1L8DFD0 A0A182M1R2 A0A1L8DF83 E2A4J1 A0A182WZA8 A0A1C7CZM0 A0A182NPD2 T1PIT2 A0A1I8NGC6 A0A182HPW0 A0A182U1D7 A0A182UQN7 A0A026W393 A0A182UBV2 A0A182FW58 A0A182SB54 W5JJV0 Q7Q024 A0A2J7QE56 A0A2M4AEH4 D6X016 A0A2M4AEJ1 A0A2M4AEY6 A0A2P8XF39 A0A2M4CN26 A0A0L0CQE9 A0A1I8NN12 A0A2M4AFX6 A0A1A9XP26 A0A1B0BNI6 E2B9R1 A0A151WLJ9 A0A1B0G7Q1 A0A1A9ZWD8 A0A1A9V8D1 A0A2M4BIF5 A0A182W9C7 A0A151IXJ2 A0A2M4BLV3 A0A195FQL9 B4PA70 B4QAZ8 A0A1I8M1P2 Q8MLQ7 W8CAE6 B3NPS7 A0A195CU57 A0A1Y1L0W9 A0A1Q3G0C6 B4I8T7 Q0IEA9 A0A0A1XJU0 F4WIX4 A0A034WQX0 A0A084WHI6 B4GBK8 A0A158N976 A0A3B0JXX9 B4LIZ5 A0A1J1ICI9 A0A0K8VSF9 B4KUC7 A0A1W4UMN4 A0A1W4V1P0 B4MYD6 B3ME82 A0A1Y1KX95 A0A1A9WIQ7 A0A0M4EV96 A0A0J7L656 B4JVU0 A0A0M9ADJ5 Q9W1J2 A0A0L7QJV6 A0A195B4K6 A0A2J7QE64 A0A182GZS3 A0A3L8DA70 A0A0A8J8P2 A0A1I8NN11 A0A067RK37 A0A1L8DDZ9 A0A1L8DDX2 A0A1Y1JT67
EC Number
2.7.11.13
Pubmed
19121390
22118469
26354079
17510324
25244985
20798317
+ More
20966253 25315136 24508170 20920257 23761445 12364791 14747013 17210077 18362917 19820115 29403074 26108605 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 28004739 25830018 21719571 25348373 24438588 21347285 26483478 30249741 24845553
20966253 25315136 24508170 20920257 23761445 12364791 14747013 17210077 18362917 19820115 29403074 26108605 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 28004739 25830018 21719571 25348373 24438588 21347285 26483478 30249741 24845553
EMBL
BABH01013656
BABH01013657
AGBW02010237
OWR48877.1
RSAL01000258
RVE43371.1
+ More
ODYU01003876 SOQ43185.1 KQ460397 KPJ15221.1 KQ459599 KPI94478.1 NWSH01004606 PCG64856.1 AXCN02001824 AJWK01009053 GFDF01009040 JAV05044.1 AXCM01006009 GFDF01009039 JAV05045.1 GL436677 EFN71650.1 KA648015 AFP62644.1 APCN01003282 KK107455 EZA50493.1 ADMH02001285 ETN63180.1 AAAB01008986 EAA00109.4 NEVH01015327 PNF26868.1 GGFK01005863 MBW39184.1 KQ971372 EFA10509.1 GGFK01005862 MBW39183.1 GGFK01006019 MBW39340.1 PYGN01002375 PSN30621.1 GGFL01002536 MBW66714.1 JRES01000049 KNC34598.1 GGFK01006355 MBW39676.1 JXJN01017413 GL446584 EFN87595.1 KQ982959 KYQ48661.1 CCAG010016890 GGFJ01003681 MBW52822.1 KQ980796 KYN12965.1 GGFJ01004833 MBW53974.1 KQ981382 KYN42219.1 CM000158 EDW92396.1 CM000362 CM002911 EDX08435.1 KMY96121.1 AE013599 BT001766 AAM68271.2 AAN71521.1 GAMC01005816 JAC00740.1 CH954179 EDV56868.1 KQ977279 KYN04185.1 GEZM01071122 JAV66030.1 GFDL01001805 JAV33240.1 CH480824 EDW57012.1 CH477764 EAT36505.1 GBXI01003031 JAD11261.1 GL888177 EGI65920.1 GAKP01002260 JAC56692.1 ATLV01023839 ATLV01023840 ATLV01023841 KE525347 KFB49680.1 CH479181 EDW31303.1 ADTU01008811 ADTU01008812 OUUW01000001 SPP75948.1 CH940648 EDW60445.1 CVRI01000043 CRK96153.1 GDHF01010510 JAI41804.1 CH933808 EDW10714.2 CH963894 EDW77125.1 CH902619 EDV35477.2 GEZM01071123 JAV66029.1 CP012524 ALC41762.1 LBMM01000577 KMQ98038.1 CH916375 EDV98078.1 KQ435692 KOX81103.1 AY058792 AAL14021.1 KQ415067 KOC58831.1 KQ976618 KYM79197.1 PNF26866.1 JXUM01002147 JXUM01002148 QOIP01000010 RLU17395.1 AB901074 BAQ02374.1 KK852575 KDR20981.1 GFDF01009416 JAV04668.1 GFDF01009415 JAV04669.1 GEZM01101870 JAV52243.1
ODYU01003876 SOQ43185.1 KQ460397 KPJ15221.1 KQ459599 KPI94478.1 NWSH01004606 PCG64856.1 AXCN02001824 AJWK01009053 GFDF01009040 JAV05044.1 AXCM01006009 GFDF01009039 JAV05045.1 GL436677 EFN71650.1 KA648015 AFP62644.1 APCN01003282 KK107455 EZA50493.1 ADMH02001285 ETN63180.1 AAAB01008986 EAA00109.4 NEVH01015327 PNF26868.1 GGFK01005863 MBW39184.1 KQ971372 EFA10509.1 GGFK01005862 MBW39183.1 GGFK01006019 MBW39340.1 PYGN01002375 PSN30621.1 GGFL01002536 MBW66714.1 JRES01000049 KNC34598.1 GGFK01006355 MBW39676.1 JXJN01017413 GL446584 EFN87595.1 KQ982959 KYQ48661.1 CCAG010016890 GGFJ01003681 MBW52822.1 KQ980796 KYN12965.1 GGFJ01004833 MBW53974.1 KQ981382 KYN42219.1 CM000158 EDW92396.1 CM000362 CM002911 EDX08435.1 KMY96121.1 AE013599 BT001766 AAM68271.2 AAN71521.1 GAMC01005816 JAC00740.1 CH954179 EDV56868.1 KQ977279 KYN04185.1 GEZM01071122 JAV66030.1 GFDL01001805 JAV33240.1 CH480824 EDW57012.1 CH477764 EAT36505.1 GBXI01003031 JAD11261.1 GL888177 EGI65920.1 GAKP01002260 JAC56692.1 ATLV01023839 ATLV01023840 ATLV01023841 KE525347 KFB49680.1 CH479181 EDW31303.1 ADTU01008811 ADTU01008812 OUUW01000001 SPP75948.1 CH940648 EDW60445.1 CVRI01000043 CRK96153.1 GDHF01010510 JAI41804.1 CH933808 EDW10714.2 CH963894 EDW77125.1 CH902619 EDV35477.2 GEZM01071123 JAV66029.1 CP012524 ALC41762.1 LBMM01000577 KMQ98038.1 CH916375 EDV98078.1 KQ435692 KOX81103.1 AY058792 AAL14021.1 KQ415067 KOC58831.1 KQ976618 KYM79197.1 PNF26866.1 JXUM01002147 JXUM01002148 QOIP01000010 RLU17395.1 AB901074 BAQ02374.1 KK852575 KDR20981.1 GFDF01009416 JAV04668.1 GFDF01009415 JAV04669.1 GEZM01101870 JAV52243.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000053240
UP000053268
UP000218220
+ More
UP000075881 UP000075880 UP000075900 UP000075886 UP000076408 UP000075885 UP000092461 UP000075883 UP000000311 UP000076407 UP000075882 UP000075884 UP000095301 UP000075840 UP000075902 UP000075903 UP000053097 UP000069272 UP000075901 UP000000673 UP000007062 UP000235965 UP000007266 UP000245037 UP000037069 UP000095300 UP000092443 UP000092460 UP000008237 UP000075809 UP000092444 UP000092445 UP000078200 UP000075920 UP000078492 UP000078541 UP000002282 UP000000304 UP000000803 UP000008711 UP000078542 UP000001292 UP000008820 UP000007755 UP000030765 UP000008744 UP000005205 UP000268350 UP000008792 UP000183832 UP000009192 UP000192221 UP000007798 UP000007801 UP000091820 UP000092553 UP000036403 UP000001070 UP000053105 UP000053825 UP000078540 UP000069940 UP000279307 UP000027135
UP000075881 UP000075880 UP000075900 UP000075886 UP000076408 UP000075885 UP000092461 UP000075883 UP000000311 UP000076407 UP000075882 UP000075884 UP000095301 UP000075840 UP000075902 UP000075903 UP000053097 UP000069272 UP000075901 UP000000673 UP000007062 UP000235965 UP000007266 UP000245037 UP000037069 UP000095300 UP000092443 UP000092460 UP000008237 UP000075809 UP000092444 UP000092445 UP000078200 UP000075920 UP000078492 UP000078541 UP000002282 UP000000304 UP000000803 UP000008711 UP000078542 UP000001292 UP000008820 UP000007755 UP000030765 UP000008744 UP000005205 UP000268350 UP000008792 UP000183832 UP000009192 UP000192221 UP000007798 UP000007801 UP000091820 UP000092553 UP000036403 UP000001070 UP000053105 UP000053825 UP000078540 UP000069940 UP000279307 UP000027135
Interpro
IPR020846
MFS_dom
+ More
IPR005829 Sugar_transporter_CS
IPR036259 MFS_trans_sf
IPR005828 MFS_sugar_transport-like
IPR004299 MBOAT_fam
IPR011009 Kinase-like_dom_sf
IPR034659 Atypical_PKC
IPR000719 Prot_kinase_dom
IPR017892 Pkinase_C
IPR020454 DAG/PE-bd
IPR000961 AGC-kinase_C
IPR008271 Ser/Thr_kinase_AS
IPR002219 PE/DAG-bd
IPR017441 Protein_kinase_ATP_BS
IPR003663 Sugar/inositol_transpt
IPR005829 Sugar_transporter_CS
IPR036259 MFS_trans_sf
IPR005828 MFS_sugar_transport-like
IPR004299 MBOAT_fam
IPR011009 Kinase-like_dom_sf
IPR034659 Atypical_PKC
IPR000719 Prot_kinase_dom
IPR017892 Pkinase_C
IPR020454 DAG/PE-bd
IPR000961 AGC-kinase_C
IPR008271 Ser/Thr_kinase_AS
IPR002219 PE/DAG-bd
IPR017441 Protein_kinase_ATP_BS
IPR003663 Sugar/inositol_transpt
ProteinModelPortal
H9J7D2
A0A212F561
A0A3S2NBT7
A0A2H1VQR6
A0A194RH13
A0A194PNB3
+ More
A0A2A4IZE6 A0A182JVB0 A0A182IM61 A0A182RGI9 A0A182QYL2 A0A1S4FT49 A0A182Y1Z9 A0A182PRA4 A0A1B0CEP4 A0A1L8DFD0 A0A182M1R2 A0A1L8DF83 E2A4J1 A0A182WZA8 A0A1C7CZM0 A0A182NPD2 T1PIT2 A0A1I8NGC6 A0A182HPW0 A0A182U1D7 A0A182UQN7 A0A026W393 A0A182UBV2 A0A182FW58 A0A182SB54 W5JJV0 Q7Q024 A0A2J7QE56 A0A2M4AEH4 D6X016 A0A2M4AEJ1 A0A2M4AEY6 A0A2P8XF39 A0A2M4CN26 A0A0L0CQE9 A0A1I8NN12 A0A2M4AFX6 A0A1A9XP26 A0A1B0BNI6 E2B9R1 A0A151WLJ9 A0A1B0G7Q1 A0A1A9ZWD8 A0A1A9V8D1 A0A2M4BIF5 A0A182W9C7 A0A151IXJ2 A0A2M4BLV3 A0A195FQL9 B4PA70 B4QAZ8 A0A1I8M1P2 Q8MLQ7 W8CAE6 B3NPS7 A0A195CU57 A0A1Y1L0W9 A0A1Q3G0C6 B4I8T7 Q0IEA9 A0A0A1XJU0 F4WIX4 A0A034WQX0 A0A084WHI6 B4GBK8 A0A158N976 A0A3B0JXX9 B4LIZ5 A0A1J1ICI9 A0A0K8VSF9 B4KUC7 A0A1W4UMN4 A0A1W4V1P0 B4MYD6 B3ME82 A0A1Y1KX95 A0A1A9WIQ7 A0A0M4EV96 A0A0J7L656 B4JVU0 A0A0M9ADJ5 Q9W1J2 A0A0L7QJV6 A0A195B4K6 A0A2J7QE64 A0A182GZS3 A0A3L8DA70 A0A0A8J8P2 A0A1I8NN11 A0A067RK37 A0A1L8DDZ9 A0A1L8DDX2 A0A1Y1JT67
A0A2A4IZE6 A0A182JVB0 A0A182IM61 A0A182RGI9 A0A182QYL2 A0A1S4FT49 A0A182Y1Z9 A0A182PRA4 A0A1B0CEP4 A0A1L8DFD0 A0A182M1R2 A0A1L8DF83 E2A4J1 A0A182WZA8 A0A1C7CZM0 A0A182NPD2 T1PIT2 A0A1I8NGC6 A0A182HPW0 A0A182U1D7 A0A182UQN7 A0A026W393 A0A182UBV2 A0A182FW58 A0A182SB54 W5JJV0 Q7Q024 A0A2J7QE56 A0A2M4AEH4 D6X016 A0A2M4AEJ1 A0A2M4AEY6 A0A2P8XF39 A0A2M4CN26 A0A0L0CQE9 A0A1I8NN12 A0A2M4AFX6 A0A1A9XP26 A0A1B0BNI6 E2B9R1 A0A151WLJ9 A0A1B0G7Q1 A0A1A9ZWD8 A0A1A9V8D1 A0A2M4BIF5 A0A182W9C7 A0A151IXJ2 A0A2M4BLV3 A0A195FQL9 B4PA70 B4QAZ8 A0A1I8M1P2 Q8MLQ7 W8CAE6 B3NPS7 A0A195CU57 A0A1Y1L0W9 A0A1Q3G0C6 B4I8T7 Q0IEA9 A0A0A1XJU0 F4WIX4 A0A034WQX0 A0A084WHI6 B4GBK8 A0A158N976 A0A3B0JXX9 B4LIZ5 A0A1J1ICI9 A0A0K8VSF9 B4KUC7 A0A1W4UMN4 A0A1W4V1P0 B4MYD6 B3ME82 A0A1Y1KX95 A0A1A9WIQ7 A0A0M4EV96 A0A0J7L656 B4JVU0 A0A0M9ADJ5 Q9W1J2 A0A0L7QJV6 A0A195B4K6 A0A2J7QE64 A0A182GZS3 A0A3L8DA70 A0A0A8J8P2 A0A1I8NN11 A0A067RK37 A0A1L8DDZ9 A0A1L8DDX2 A0A1Y1JT67
PDB
4LDS
E-value=5.09738e-21,
Score=250
Ontologies
GO
Topology
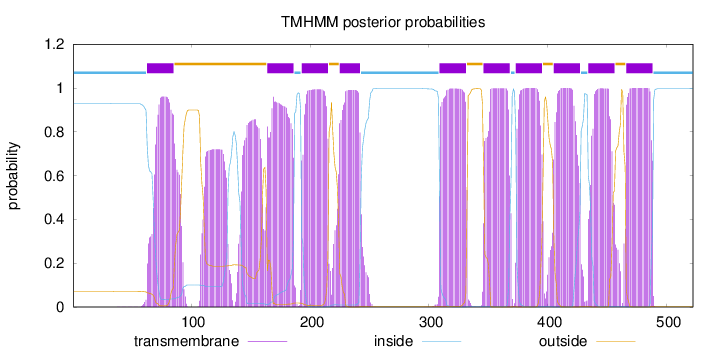
Length:
522
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
249.69816
Exp number, first 60 AAs:
0.02821
Total prob of N-in:
0.92861
inside
1 - 62
TMhelix
63 - 85
outside
86 - 163
TMhelix
164 - 186
inside
187 - 192
TMhelix
193 - 215
outside
216 - 224
TMhelix
225 - 242
inside
243 - 308
TMhelix
309 - 331
outside
332 - 345
TMhelix
346 - 368
inside
369 - 372
TMhelix
373 - 395
outside
396 - 404
TMhelix
405 - 427
inside
428 - 433
TMhelix
434 - 456
outside
457 - 465
TMhelix
466 - 488
inside
489 - 522
Population Genetic Test Statistics
Pi
267.269352
Theta
178.407289
Tajima's D
1.40482
CLR
0.127666
CSRT
0.766261686915654
Interpretation
Uncertain
