Gene
KWMTBOMO04447
Pre Gene Modal
BGIBMGA005419
Annotation
PREDICTED:_mucin-2-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.046
Sequence
CDS
ATGATGAATTTTTTTAGTAAAGCTCCAACCGTGAAGGAAATTCAAAGGCAAAATGACCGTGATCTACGAAAGGCTGGTCGCGACCTGGAAAGAGATAAAGCTAACCTGGAAAGGGAAGAAAAGAAATTAGTAAAGTGGTGTTTCGCTGCAGAAAATTTTTTAGTACTCGAAGGCATGAAGCATTGCATCAAGCCAGAACCGGGGAAAGAAATAGCAAGCGCGGATGACGAAAAAACTAAAGCCAAATTGATCATGACCATCGACCCGTCGTTGTACTGTCCGAGTTTAGAAGCCGAAAAAAGAAAACAATCCAACGCCTTTAGTGCTATTTTCTTAAGTGGAAATTTCAACTGCGATAGTTGGTACATAGATAGCGGGGCCAGCAAACACATGACAGCAAAGAAAGAATACGTTGAGAGTCCATCATTTGAACATGAAATTAAAGAAATAATTGTTGCGAACAAAACATCCGTCCCGGTAGTGTGTTCTGGTGATGTAAACATAATCACAGTTGTGAATGGTGTAGAATATGACATCACAGTTTCAAATGTATTGTGCATACCGAGTTTAACAACAAACTTGCTTTCAGTGAGTCAGTTAATTGAGAATGGCAATAAGGTAATCTTTAAAGAGAAATTTTGCTATATTTACAATCAACAGAGAACTCTAGTTGGAAAGGCAGAATTAGTAGATGGTGTGTACAGATTATATACTAAGAAATCAGAACAGTTATTCGCAGCCACAGCTAAGGTGTCGAGTGAAACTTGGCACCGACGATTAGGGCATATCAACAGTACCAGCTTGAACAAAATGAAGAATGGAGCTACGACAGGTATTTCATACACAAATAAAGCTGATGTTGACAAATCTAAGTGTACTGTTTGCTGTAAGGGGAAGCAGTCTAGACTACCTTTTCAGCCTAGTTCAACTAACACCCAAGATATTTTAGAATTAATACACTCAGATGTGTGTGGTCCGATGGAAAATGTGTCCATTGGAGGATCCAGATACTATGTCTTATTTGTGGATGACTACAGCAGAATGGTTTTTGTATACTTCATGAAAACAAAGTCAGAGGTATTTAAGTTTTTCAAAGAATTCCAGTCACTTGTAGAAAAACAGACAGATCGGAAAATTAAAATTCTAAGAACAGACAATGGCGGTGAGTACTGCAGTCAAGACTTTGAAAGATATCTAAAACAACACGGAATAATTCACCAGAAAAGTAATGCTTACACACCACAGCAAAATGGTGTGTGTGAAAGGATGAATAGGTCACTAGTTGAAAAGGCTAGATGTCTAATTTTTGATGCTGGTTTACATAAAAAGTTTTGGGCAGAAGCCATAAATACATCTGTATACTTAAGGAATCGATCAGTTGTTACTGGATTAAACAACATGACTCCATATGAAGCCTGGACAAAGAAAAAACCTGACTTAAGTCATGTACGTATTTTTGGAAGTAAGGTTATGATGCACATACCAAAAGAAAAGCGCTTAAAATGGGATATGAAGGCTAAAGAACTTATTTTGGTAGGCTATGCAGACAATGTAAAAGGCTATAGGGTTTATAACCCAGAAAATAACTCTGTCACAACTAGCAGAGACATTATAATAATGGAGAAAACTGAAAATACAATTGAAATTCCTATTGAGTGCAAGGCTCCAGCGGAGGAACAGAGAGAAGGAGACGAAAGTAGAACTACAGAAAGTACAGACACTGAATGTGATGATAATGATATCACATATATTCCAGAAGCTTCTTCAGGAAGTGACTCATATGACAGTTTTTCTGAAACACCATCAGAACCAGAACCTTCATTATTAGAATTGAATCAGGATATTCCAAACAAGCGAGTGAGAAAAATACCTGATAGATTTAGCTATAGTCACTTATGTATTGCAGAATCAGATGATCAAAATATAATGGATATTTCACTCAAGGAAGCTTTAAATGGACCTGAAAGTAAATTTTGGAGATCAAGCATGGAGGAGGAATTAGAATCATTTAAGAAAAATGATGCATGGGAACTTGTGGATAAGCCTCGTGACTCTACAGTAGTGCAGTGCAAATGGGTATTTAAAAAGAAGTATGATAGTGTCGGTAATGTGCGTTATCGTGCTAGATTAGTGGCAAAAGGCTTTTCACAGAAAGCTGGCATAGACTATCATGAAACTTTTTCACCTGTGCTACGTTATACTACTTTAAGATTGTTATTTTCTATAGCAGTAAAACTAGGTTTAGATATACGTCATCTTGATGTCACTACAGCCTTTCTAAATGGTTTTTTGAATGAATCTGTGTATATGCAAAAACCTGTCCTTTATAGAGATTGTAATGATAATGATAACAAAGTATTAAAGCTTAAACGTGCCATATATGGACTTAAGCAATCATCTCGTGCTTGGTATCAGAGAGTTAATGATTACTTAGAGTCTTTAGGATATTTAAAATCTAAGTATGAACCGTGTTTGTTTACGAAGTCTGAAGGAAATGCTAAAGTTATAATAGCATTGTTTGTTGATGATTTTTTTGTTTTTTCTAACTGTAAAATGGCAACTAATCAGTTAATACAAGATTTGAGTAGTAAATTTAAAATTAAAGATTTGGGTCAAATAAAGCAATGTTTAGGCTTAAGAGTGAATATGTATAAAAATGCTATAACTGTTGATCAAGAACAGTATATAGAAAGCTTGTTAAAAAAGTTTAACATGTCACATTGTAAAACTATAGAAACTCCTATGGAGATTAACTTAAGATTGGAAAAATCTGAGAATAGTTTATGTAATAAAAATATTCCATATAGACAATTAATAGGTAACTTAATGTATTTAGCTGTGTTAACCAGACCAGACATAGCTTACAGTGAAATGGAAATAAAGAAAATGGCTAAAGAAGGTAACAATGATGGTTGCAAAATATTAGCCAAACAATTAGTTCAATTAAGAAAACAGAAAAACAGAATTTATGCTGCAAATAGCAAGATATCAAGTGTACAGATCCACAATAAAGCAATGGGTGCAAATATTGCTATTGCAGGAGCAATGGGCACTACAGCTAAAACAATGGGCAACATGAATAAGGTTATGAATCCACATCAAATTGCAAAGGATATGGAGGCATTCAAACAGGCAAATGCCAAAATGGATATGACAGATGAAATGATTTCAGACACACTAGATGATATCATGGACGAATCTGGTGATGAGGAAGAATCTGCTGGAATTGTCAACCAGGTGCTGGATGAGATCGGTATAGAAATCAGCGGGAAGATGGCAGACGCTCCTTCAGTTTCCCGGAACAAAATCGGACAATCAACTACAGACGCCGACAAAGATATAATGGCACAACTCGCTAAACTGAAATCCACATAG
Protein
MMNFFSKAPTVKEIQRQNDRDLRKAGRDLERDKANLEREEKKLVKWCFAAENFLVLEGMKHCIKPEPGKEIASADDEKTKAKLIMTIDPSLYCPSLEAEKRKQSNAFSAIFLSGNFNCDSWYIDSGASKHMTAKKEYVESPSFEHEIKEIIVANKTSVPVVCSGDVNIITVVNGVEYDITVSNVLCIPSLTTNLLSVSQLIENGNKVIFKEKFCYIYNQQRTLVGKAELVDGVYRLYTKKSEQLFAATAKVSSETWHRRLGHINSTSLNKMKNGATTGISYTNKADVDKSKCTVCCKGKQSRLPFQPSSTNTQDILELIHSDVCGPMENVSIGGSRYYVLFVDDYSRMVFVYFMKTKSEVFKFFKEFQSLVEKQTDRKIKILRTDNGGEYCSQDFERYLKQHGIIHQKSNAYTPQQNGVCERMNRSLVEKARCLIFDAGLHKKFWAEAINTSVYLRNRSVVTGLNNMTPYEAWTKKKPDLSHVRIFGSKVMMHIPKEKRLKWDMKAKELILVGYADNVKGYRVYNPENNSVTTSRDIIIMEKTENTIEIPIECKAPAEEQREGDESRTTESTDTECDDNDITYIPEASSGSDSYDSFSETPSEPEPSLLELNQDIPNKRVRKIPDRFSYSHLCIAESDDQNIMDISLKEALNGPESKFWRSSMEEELESFKKNDAWELVDKPRDSTVVQCKWVFKKKYDSVGNVRYRARLVAKGFSQKAGIDYHETFSPVLRYTTLRLLFSIAVKLGLDIRHLDVTTAFLNGFLNESVYMQKPVLYRDCNDNDNKVLKLKRAIYGLKQSSRAWYQRVNDYLESLGYLKSKYEPCLFTKSEGNAKVIIALFVDDFFVFSNCKMATNQLIQDLSSKFKIKDLGQIKQCLGLRVNMYKNAITVDQEQYIESLLKKFNMSHCKTIETPMEINLRLEKSENSLCNKNIPYRQLIGNLMYLAVLTRPDIAYSEMEIKKMAKEGNNDGCKILAKQLVQLRKQKNRIYAANSKISSVQIHNKAMGANIAIAGAMGTTAKTMGNMNKVMNPHQIAKDMEAFKQANAKMDMTDEMISDTLDDIMDESGDEEESAGIVNQVLDEIGIEISGKMADAPSVSRNKIGQSTTDADKDIMAQLAKLKST
Summary
Uniprot
A0A2A4J959
A0A0N1ICY1
A0A194RU38
A0A1Y1LYU4
A0A2J7RKJ6
A0A2J7R8S8
+ More
A0A2J7R668 A0A2J7Q1Z2 A0A146LH62 A0A0A9WRE3 A0A251T6N3 A0A2J7PSB2 A0A085MVP5 A0A085N572 A0A0K8U6S2 A0A085NQR3 A0A085MSF4 A0A085N9K0 A0A0A9WPH5 A0A3S3NIE3 A0A151T469 A0A151QNH5 A0A293MM55 K7J8U2 A0A0J7K6W1 A0A151R7U0 A0A151TMN9 A0A151T355 A0A0K8WJW9 A0A0K8U6X2 A0A151TIH6 A0A0A1WFC7 A0A0V0RWJ3 A0A2N9IWX1 A0A224X581 A0A151THM1 B6REL8 A0A085MV21 A0A0V1MGJ0
A0A2J7R668 A0A2J7Q1Z2 A0A146LH62 A0A0A9WRE3 A0A251T6N3 A0A2J7PSB2 A0A085MVP5 A0A085N572 A0A0K8U6S2 A0A085NQR3 A0A085MSF4 A0A085N9K0 A0A0A9WPH5 A0A3S3NIE3 A0A151T469 A0A151QNH5 A0A293MM55 K7J8U2 A0A0J7K6W1 A0A151R7U0 A0A151TMN9 A0A151T355 A0A0K8WJW9 A0A0K8U6X2 A0A151TIH6 A0A0A1WFC7 A0A0V0RWJ3 A0A2N9IWX1 A0A224X581 A0A151THM1 B6REL8 A0A085MV21 A0A0V1MGJ0
EMBL
NWSH01002292
PCG68657.1
PCG68658.1
KQ460882
KPJ11482.1
KQ459875
+ More
KPJ19636.1 GEZM01045163 GEZM01045162 JAV77888.1 NEVH01002725 PNF41351.1 NEVH01006721 PNF37230.1 NEVH01006980 PNF36330.1 NEVH01019375 PNF22597.1 GDHC01011760 JAQ06869.1 GBHO01033235 JAG10369.1 CM007900 OTG06758.1 NEVH01021933 PNF19227.1 KL367628 KFD61291.1 KL367553 KFD64618.1 GDHF01030254 JAI22060.1 KL367480 KFD71809.1 KL367691 KFD60150.1 KL367527 KFD66146.1 GBHO01033237 JAG10367.1 NCKU01012701 RWS00004.1 CM003610 KYP61818.1 KYP63036.1 KQ485638 KYP31826.1 GFWV01017285 MAA42013.1 AAZX01000499 LBMM01012645 KMQ86082.1 KQ483989 KYP38573.1 CM003606 KYP68287.1 KYP61490.1 GDHF01000873 JAI51441.1 GDHF01029905 JAI22409.1 CM003608 KYP66838.1 GBXI01017107 JAC97184.1 JYDL01000068 KRX18751.1 OIVN01006239 SPD28653.1 GFTR01008771 JAW07655.1 KYP66486.1 EU180847 ABW74566.1 KL367637 KFD61067.1 JYDO01000104 KRZ71003.1
KPJ19636.1 GEZM01045163 GEZM01045162 JAV77888.1 NEVH01002725 PNF41351.1 NEVH01006721 PNF37230.1 NEVH01006980 PNF36330.1 NEVH01019375 PNF22597.1 GDHC01011760 JAQ06869.1 GBHO01033235 JAG10369.1 CM007900 OTG06758.1 NEVH01021933 PNF19227.1 KL367628 KFD61291.1 KL367553 KFD64618.1 GDHF01030254 JAI22060.1 KL367480 KFD71809.1 KL367691 KFD60150.1 KL367527 KFD66146.1 GBHO01033237 JAG10367.1 NCKU01012701 RWS00004.1 CM003610 KYP61818.1 KYP63036.1 KQ485638 KYP31826.1 GFWV01017285 MAA42013.1 AAZX01000499 LBMM01012645 KMQ86082.1 KQ483989 KYP38573.1 CM003606 KYP68287.1 KYP61490.1 GDHF01000873 JAI51441.1 GDHF01029905 JAI22409.1 CM003608 KYP66838.1 GBXI01017107 JAC97184.1 JYDL01000068 KRX18751.1 OIVN01006239 SPD28653.1 GFTR01008771 JAW07655.1 KYP66486.1 EU180847 ABW74566.1 KL367637 KFD61067.1 JYDO01000104 KRZ71003.1
Proteomes
Pfam
Interpro
IPR039537
Retrotran_Ty1/copia-like
+ More
IPR001584 Integrase_cat-core
IPR036875 Znf_CCHC_sf
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR013103 RVT_2
IPR025724 GAG-pre-integrase_dom
IPR025314 DUF4219
IPR036397 RNaseH_sf
IPR009056 Cyt_c-like_dom
IPR008042 Retrotrans_Pao
IPR000477 RT_dom
IPR008737 Peptidase_asp_put
IPR001584 Integrase_cat-core
IPR036875 Znf_CCHC_sf
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR013103 RVT_2
IPR025724 GAG-pre-integrase_dom
IPR025314 DUF4219
IPR036397 RNaseH_sf
IPR009056 Cyt_c-like_dom
IPR008042 Retrotrans_Pao
IPR000477 RT_dom
IPR008737 Peptidase_asp_put
Gene 3D
ProteinModelPortal
A0A2A4J959
A0A0N1ICY1
A0A194RU38
A0A1Y1LYU4
A0A2J7RKJ6
A0A2J7R8S8
+ More
A0A2J7R668 A0A2J7Q1Z2 A0A146LH62 A0A0A9WRE3 A0A251T6N3 A0A2J7PSB2 A0A085MVP5 A0A085N572 A0A0K8U6S2 A0A085NQR3 A0A085MSF4 A0A085N9K0 A0A0A9WPH5 A0A3S3NIE3 A0A151T469 A0A151QNH5 A0A293MM55 K7J8U2 A0A0J7K6W1 A0A151R7U0 A0A151TMN9 A0A151T355 A0A0K8WJW9 A0A0K8U6X2 A0A151TIH6 A0A0A1WFC7 A0A0V0RWJ3 A0A2N9IWX1 A0A224X581 A0A151THM1 B6REL8 A0A085MV21 A0A0V1MGJ0
A0A2J7R668 A0A2J7Q1Z2 A0A146LH62 A0A0A9WRE3 A0A251T6N3 A0A2J7PSB2 A0A085MVP5 A0A085N572 A0A0K8U6S2 A0A085NQR3 A0A085MSF4 A0A085N9K0 A0A0A9WPH5 A0A3S3NIE3 A0A151T469 A0A151QNH5 A0A293MM55 K7J8U2 A0A0J7K6W1 A0A151R7U0 A0A151TMN9 A0A151T355 A0A0K8WJW9 A0A0K8U6X2 A0A151TIH6 A0A0A1WFC7 A0A0V0RWJ3 A0A2N9IWX1 A0A224X581 A0A151THM1 B6REL8 A0A085MV21 A0A0V1MGJ0
PDB
3FRT
E-value=0.000135288,
Score=112
Ontologies
GO
PANTHER
Topology
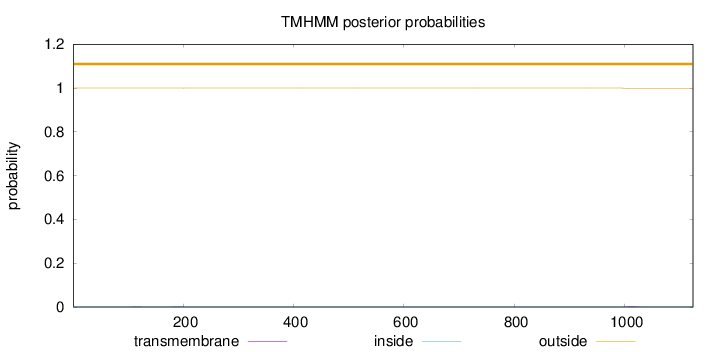
Length:
1124
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04557
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00037
outside
1 - 1124
Population Genetic Test Statistics
Pi
431.268556
Theta
197.344269
Tajima's D
2.740929
CLR
0.000392
CSRT
0.96285185740713
Interpretation
Possibly Balancing Selection
