Gene
KWMTBOMO04441 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005380
Annotation
PREDICTED:_venom_protease-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.393
Sequence
CDS
ATGAGGCAATATGGTCCTAAGCATTTAATATGGATATTAATTATATCCGGCATTCTTATAAAAGCCGAAGACGTTGGAGATGAATGCACCCCAAATAACCAGATATCTGAAGGTATCTGTACTCTGGTGAATGATTGTCCTCAGGCTGTAATGGCAATAAAAAATAAAAGGTTTCAACCATTTCAAAGATGTGGATTCAGAGGATTTCAGGAGATAGTTTGTTGCCCTACAACTGTGGACAAATTTGGTCAAACAGAGAAAACAAGAACTATTAAACGAATTGCTGAACGAGAATGTGACAAAATCATATCTTCAACTGTACCTCCGCTCGACCTGTATATACTTGGCGGGGAAGCTGCATCGATGGGAGAATTTCCTTACATGGTTGCTGTTGGCTTCGACCGTGGTAATGGTTACGAATTCGATTGCGGAGGTTCGTTGCTATCCAACCTGTATGTACTGACAGCTGCGCACTGCGTCGACACATTGGATCGGATAGAGCCCAGTTTGGTGCGCGTCGGTGTTATAGAGCTGGGCAGCAACGCTTGGACCGAGGGCACGGACTACCGCATCGCGCAAATCATAACCCACCCGAATTACACCAGACGAGAGAAATACCATGACTTGGCCTTACTCAGGTTGGAAAAAGCAGTCGCATTCTCGAGCAACGTAAATGCTGTGTGTTTGGAAACGAGTACAGAAGATCCAACTGTTCCATTGACCATCACCGGTTGGGGGAAGATAAGTAATACTAGGAACGCTAAAAGCAATATATTACTCAAGGCGAATGTGACCGCTGTAAAGGCTGAGAAGTGTAGTGAATCCTACCTGAATTGGCGTAAACTACCTAAAGGCATATCCGACAGTCAGATTTGTGCTGGAGATCCTGAAGGCATCAGGGATACTTGTCAGGGCGACTCGGGCGGTCCGCTGCAGATGTGGCGCGACGTGTACCGGCTAGTCGGCGTGACGTCATTCGGGCGCGGCTGCGGCTCGCCCGTGCCCGGCGTGTACACGCGCCTGTCGCGCTACCTCGACTGGATCGAATCTGTCGTATGGCCCAATCACGTTTAA
Protein
MRQYGPKHLIWILIISGILIKAEDVGDECTPNNQISEGICTLVNDCPQAVMAIKNKRFQPFQRCGFRGFQEIVCCPTTVDKFGQTEKTRTIKRIAERECDKIISSTVPPLDLYILGGEAASMGEFPYMVAVGFDRGNGYEFDCGGSLLSNLYVLTAAHCVDTLDRIEPSLVRVGVIELGSNAWTEGTDYRIAQIITHPNYTRREKYHDLALLRLEKAVAFSSNVNAVCLETSTEDPTVPLTITGWGKISNTRNAKSNILLKANVTAVKAEKCSESYLNWRKLPKGISDSQICAGDPEGIRDTCQGDSGGPLQMWRDVYRLVGVTSFGRGCGSPVPGVYTRLSRYLDWIESVVWPNHV
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
A0A290U6G2
A0A1E1WR09
Q5MPC8
A0A2H1WH67
A0A194PXG1
S4PBS4
+ More
A0A191XQK8 A0A212FEM3 A0A194RCA3 A0A3S2M5Y0 A0A212FFV4 A0A034VK88 A0A0K8TZ88 A0A1V1FKN1 A0A1Y1ND16 A0A1W4WG82 D6WV12 A0A2J7QCI9 T1PLT8 W8BWP2 A0A2P8XRL8 A0A1Q3FGD5 A0A0T6AT77 B0X814 Q9XY63 A0A1J1INC2 A0A182M4M7 A0A2M4BSS9 A0A182UFN5 A7USI2 W5JTH3 A0A2C9GS33 A0A182KF51 A0A2M4AVR2 A0A182VBQ0 A0A2M4AVR1 A0A0K8VST9 T1E9T1 A0A0A1XH26 A0A034WJS9 A0A182J5D5 A0A084VFL8 A0A2M4BUS3 A0A182PZZ7 A0A0L7R7Y2 B3MWJ3 Q17CN4 A0A195CK42 A0A026WZH2 T1PES6 A0A1I8NHA9 A0A1I8NHA4 D3TMC3 A0A3L8DCB2 E2ARP5 A0A1A9Y007 A0A026WCC0 A0A0M9AB69 Q171M7 A0A1Y1N017 A0A0P6ISZ8 A0A088ATI0 W8C4A8 A0A348G5Y6 D6WCU1 A0A1W6EW85 A0A2A3EIB4 A0A195EVD6 F4X254 A0A1S4FH91 A0A2M4DQF4 A0A1Y1KPC1 J9HYX9 Q16NE9 A0A0A1XND1 N6TSF6 A0A0A1WWW1 A0A182VFE5 A0A182KNG1 Q5TNC5 W5JRY9 A0A2M4AP36 A0A1I8P9N3 A0A1B0D6M3 A0A1B0FMC8 A0A0P6IUV8 A0A182X8T2 A0A1S4FGZ3 B0W6K2 A0A0A1XBJ5 A0A023ESS7 A0A182RV45
A0A191XQK8 A0A212FEM3 A0A194RCA3 A0A3S2M5Y0 A0A212FFV4 A0A034VK88 A0A0K8TZ88 A0A1V1FKN1 A0A1Y1ND16 A0A1W4WG82 D6WV12 A0A2J7QCI9 T1PLT8 W8BWP2 A0A2P8XRL8 A0A1Q3FGD5 A0A0T6AT77 B0X814 Q9XY63 A0A1J1INC2 A0A182M4M7 A0A2M4BSS9 A0A182UFN5 A7USI2 W5JTH3 A0A2C9GS33 A0A182KF51 A0A2M4AVR2 A0A182VBQ0 A0A2M4AVR1 A0A0K8VST9 T1E9T1 A0A0A1XH26 A0A034WJS9 A0A182J5D5 A0A084VFL8 A0A2M4BUS3 A0A182PZZ7 A0A0L7R7Y2 B3MWJ3 Q17CN4 A0A195CK42 A0A026WZH2 T1PES6 A0A1I8NHA9 A0A1I8NHA4 D3TMC3 A0A3L8DCB2 E2ARP5 A0A1A9Y007 A0A026WCC0 A0A0M9AB69 Q171M7 A0A1Y1N017 A0A0P6ISZ8 A0A088ATI0 W8C4A8 A0A348G5Y6 D6WCU1 A0A1W6EW85 A0A2A3EIB4 A0A195EVD6 F4X254 A0A1S4FH91 A0A2M4DQF4 A0A1Y1KPC1 J9HYX9 Q16NE9 A0A0A1XND1 N6TSF6 A0A0A1WWW1 A0A182VFE5 A0A182KNG1 Q5TNC5 W5JRY9 A0A2M4AP36 A0A1I8P9N3 A0A1B0D6M3 A0A1B0FMC8 A0A0P6IUV8 A0A182X8T2 A0A1S4FGZ3 B0W6K2 A0A0A1XBJ5 A0A023ESS7 A0A182RV45
Pubmed
EMBL
KY680240
ATD13321.1
GDQN01001650
JAT89404.1
AY672782
AAV91004.1
+ More
ODYU01008646 SOQ52408.1 KQ459589 KPI97708.1 GAIX01004461 JAA88099.1 KU360127 ANJ42865.1 AGBW02008921 OWR52184.1 KQ460397 KPJ15232.1 RSAL01000025 RVE52129.1 AGBW02008764 OWR52603.1 GAKP01016974 GAKP01016970 JAC41978.1 GDHF01032716 GDHF01010346 JAI19598.1 JAI41968.1 FX985359 BAX07372.1 GEZM01008423 JAV94790.1 KQ971357 EFA09207.2 NEVH01016289 PNF26300.1 KA649747 AFP64376.1 GAMC01005232 JAC01324.1 PYGN01001468 PSN34643.1 GFDL01008432 JAV26613.1 LJIG01022856 KRT78368.1 DS232472 EDS42283.1 AF053921 AAD21841.1 CVRI01000054 CRK99977.1 AXCM01000582 GGFJ01006840 MBW55981.1 AAAB01008846 EDO64345.2 ADMH02000122 ETN67672.1 APCN01001124 GGFK01011556 MBW44877.1 GGFK01011555 MBW44876.1 GDHF01028047 GDHF01010406 JAI24267.1 JAI41908.1 GAMD01001613 JAA99977.1 GBXI01004036 JAD10256.1 GAKP01003131 JAC55821.1 ATLV01012446 KE524793 KFB36762.1 GGFJ01007674 MBW56815.1 AXCN02002154 KQ414637 KOC66990.1 CH902625 EDV34978.2 KPU75377.1 CH477307 EAT44063.1 KQ977642 KYN01091.1 KK107077 EZA60544.1 KA647241 AFP61870.1 EZ422575 ADD18851.1 QOIP01000010 RLU17773.1 GL442157 EFN63907.1 KK107321 QOIP01000002 EZA52684.1 RLU25863.1 KQ435694 KOX80910.1 CH477450 EAT40694.2 GEZM01021578 JAV88927.1 GDUN01001005 JAN94914.1 GAMC01002402 JAC04154.1 FX985524 BBF97859.1 KQ971311 EEZ99357.2 KY563546 ARK19955.1 KZ288253 PBC30926.1 KQ981954 KYN32225.1 GL888565 EGI59470.1 GGFL01015607 MBW79785.1 GEZM01077598 JAV63253.1 EJY57695.1 CH477825 EAT35883.2 GBXI01001825 JAD12467.1 APGK01057050 KB741277 ENN71261.1 GBXI01011394 JAD02898.1 AAAB01008984 EAL38946.3 ADMH02000599 ETN65690.1 GGFK01009233 MBW42554.1 AJVK01012090 AJVK01012091 CCAG010000991 GDUN01001000 JAN94919.1 DS231848 EDS36676.1 GBXI01005976 JAD08316.1 GAPW01001752 JAC11846.1
ODYU01008646 SOQ52408.1 KQ459589 KPI97708.1 GAIX01004461 JAA88099.1 KU360127 ANJ42865.1 AGBW02008921 OWR52184.1 KQ460397 KPJ15232.1 RSAL01000025 RVE52129.1 AGBW02008764 OWR52603.1 GAKP01016974 GAKP01016970 JAC41978.1 GDHF01032716 GDHF01010346 JAI19598.1 JAI41968.1 FX985359 BAX07372.1 GEZM01008423 JAV94790.1 KQ971357 EFA09207.2 NEVH01016289 PNF26300.1 KA649747 AFP64376.1 GAMC01005232 JAC01324.1 PYGN01001468 PSN34643.1 GFDL01008432 JAV26613.1 LJIG01022856 KRT78368.1 DS232472 EDS42283.1 AF053921 AAD21841.1 CVRI01000054 CRK99977.1 AXCM01000582 GGFJ01006840 MBW55981.1 AAAB01008846 EDO64345.2 ADMH02000122 ETN67672.1 APCN01001124 GGFK01011556 MBW44877.1 GGFK01011555 MBW44876.1 GDHF01028047 GDHF01010406 JAI24267.1 JAI41908.1 GAMD01001613 JAA99977.1 GBXI01004036 JAD10256.1 GAKP01003131 JAC55821.1 ATLV01012446 KE524793 KFB36762.1 GGFJ01007674 MBW56815.1 AXCN02002154 KQ414637 KOC66990.1 CH902625 EDV34978.2 KPU75377.1 CH477307 EAT44063.1 KQ977642 KYN01091.1 KK107077 EZA60544.1 KA647241 AFP61870.1 EZ422575 ADD18851.1 QOIP01000010 RLU17773.1 GL442157 EFN63907.1 KK107321 QOIP01000002 EZA52684.1 RLU25863.1 KQ435694 KOX80910.1 CH477450 EAT40694.2 GEZM01021578 JAV88927.1 GDUN01001005 JAN94914.1 GAMC01002402 JAC04154.1 FX985524 BBF97859.1 KQ971311 EEZ99357.2 KY563546 ARK19955.1 KZ288253 PBC30926.1 KQ981954 KYN32225.1 GL888565 EGI59470.1 GGFL01015607 MBW79785.1 GEZM01077598 JAV63253.1 EJY57695.1 CH477825 EAT35883.2 GBXI01001825 JAD12467.1 APGK01057050 KB741277 ENN71261.1 GBXI01011394 JAD02898.1 AAAB01008984 EAL38946.3 ADMH02000599 ETN65690.1 GGFK01009233 MBW42554.1 AJVK01012090 AJVK01012091 CCAG010000991 GDUN01001000 JAN94919.1 DS231848 EDS36676.1 GBXI01005976 JAD08316.1 GAPW01001752 JAC11846.1
Proteomes
UP000053268
UP000007151
UP000053240
UP000283053
UP000192223
UP000007266
+ More
UP000235965 UP000245037 UP000002320 UP000183832 UP000075883 UP000075902 UP000007062 UP000000673 UP000075840 UP000075881 UP000075903 UP000075880 UP000030765 UP000075886 UP000053825 UP000007801 UP000008820 UP000078542 UP000053097 UP000095301 UP000279307 UP000000311 UP000092443 UP000053105 UP000005203 UP000242457 UP000078541 UP000007755 UP000019118 UP000075882 UP000095300 UP000092462 UP000092444 UP000076407 UP000075900
UP000235965 UP000245037 UP000002320 UP000183832 UP000075883 UP000075902 UP000007062 UP000000673 UP000075840 UP000075881 UP000075903 UP000075880 UP000030765 UP000075886 UP000053825 UP000007801 UP000008820 UP000078542 UP000053097 UP000095301 UP000279307 UP000000311 UP000092443 UP000053105 UP000005203 UP000242457 UP000078541 UP000007755 UP000019118 UP000075882 UP000095300 UP000092462 UP000092444 UP000076407 UP000075900
PRIDE
Interpro
SUPFAM
SSF50494
SSF50494
Gene 3D
CDD
ProteinModelPortal
A0A290U6G2
A0A1E1WR09
Q5MPC8
A0A2H1WH67
A0A194PXG1
S4PBS4
+ More
A0A191XQK8 A0A212FEM3 A0A194RCA3 A0A3S2M5Y0 A0A212FFV4 A0A034VK88 A0A0K8TZ88 A0A1V1FKN1 A0A1Y1ND16 A0A1W4WG82 D6WV12 A0A2J7QCI9 T1PLT8 W8BWP2 A0A2P8XRL8 A0A1Q3FGD5 A0A0T6AT77 B0X814 Q9XY63 A0A1J1INC2 A0A182M4M7 A0A2M4BSS9 A0A182UFN5 A7USI2 W5JTH3 A0A2C9GS33 A0A182KF51 A0A2M4AVR2 A0A182VBQ0 A0A2M4AVR1 A0A0K8VST9 T1E9T1 A0A0A1XH26 A0A034WJS9 A0A182J5D5 A0A084VFL8 A0A2M4BUS3 A0A182PZZ7 A0A0L7R7Y2 B3MWJ3 Q17CN4 A0A195CK42 A0A026WZH2 T1PES6 A0A1I8NHA9 A0A1I8NHA4 D3TMC3 A0A3L8DCB2 E2ARP5 A0A1A9Y007 A0A026WCC0 A0A0M9AB69 Q171M7 A0A1Y1N017 A0A0P6ISZ8 A0A088ATI0 W8C4A8 A0A348G5Y6 D6WCU1 A0A1W6EW85 A0A2A3EIB4 A0A195EVD6 F4X254 A0A1S4FH91 A0A2M4DQF4 A0A1Y1KPC1 J9HYX9 Q16NE9 A0A0A1XND1 N6TSF6 A0A0A1WWW1 A0A182VFE5 A0A182KNG1 Q5TNC5 W5JRY9 A0A2M4AP36 A0A1I8P9N3 A0A1B0D6M3 A0A1B0FMC8 A0A0P6IUV8 A0A182X8T2 A0A1S4FGZ3 B0W6K2 A0A0A1XBJ5 A0A023ESS7 A0A182RV45
A0A191XQK8 A0A212FEM3 A0A194RCA3 A0A3S2M5Y0 A0A212FFV4 A0A034VK88 A0A0K8TZ88 A0A1V1FKN1 A0A1Y1ND16 A0A1W4WG82 D6WV12 A0A2J7QCI9 T1PLT8 W8BWP2 A0A2P8XRL8 A0A1Q3FGD5 A0A0T6AT77 B0X814 Q9XY63 A0A1J1INC2 A0A182M4M7 A0A2M4BSS9 A0A182UFN5 A7USI2 W5JTH3 A0A2C9GS33 A0A182KF51 A0A2M4AVR2 A0A182VBQ0 A0A2M4AVR1 A0A0K8VST9 T1E9T1 A0A0A1XH26 A0A034WJS9 A0A182J5D5 A0A084VFL8 A0A2M4BUS3 A0A182PZZ7 A0A0L7R7Y2 B3MWJ3 Q17CN4 A0A195CK42 A0A026WZH2 T1PES6 A0A1I8NHA9 A0A1I8NHA4 D3TMC3 A0A3L8DCB2 E2ARP5 A0A1A9Y007 A0A026WCC0 A0A0M9AB69 Q171M7 A0A1Y1N017 A0A0P6ISZ8 A0A088ATI0 W8C4A8 A0A348G5Y6 D6WCU1 A0A1W6EW85 A0A2A3EIB4 A0A195EVD6 F4X254 A0A1S4FH91 A0A2M4DQF4 A0A1Y1KPC1 J9HYX9 Q16NE9 A0A0A1XND1 N6TSF6 A0A0A1WWW1 A0A182VFE5 A0A182KNG1 Q5TNC5 W5JRY9 A0A2M4AP36 A0A1I8P9N3 A0A1B0D6M3 A0A1B0FMC8 A0A0P6IUV8 A0A182X8T2 A0A1S4FGZ3 B0W6K2 A0A0A1XBJ5 A0A023ESS7 A0A182RV45
PDB
6O1G
E-value=3.55988e-35,
Score=371
Ontologies
GO
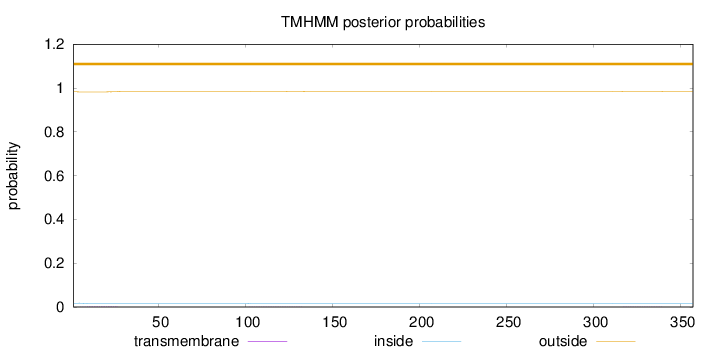
Topology
SignalP
Position: 1 - 22,
Likelihood: 0.994993
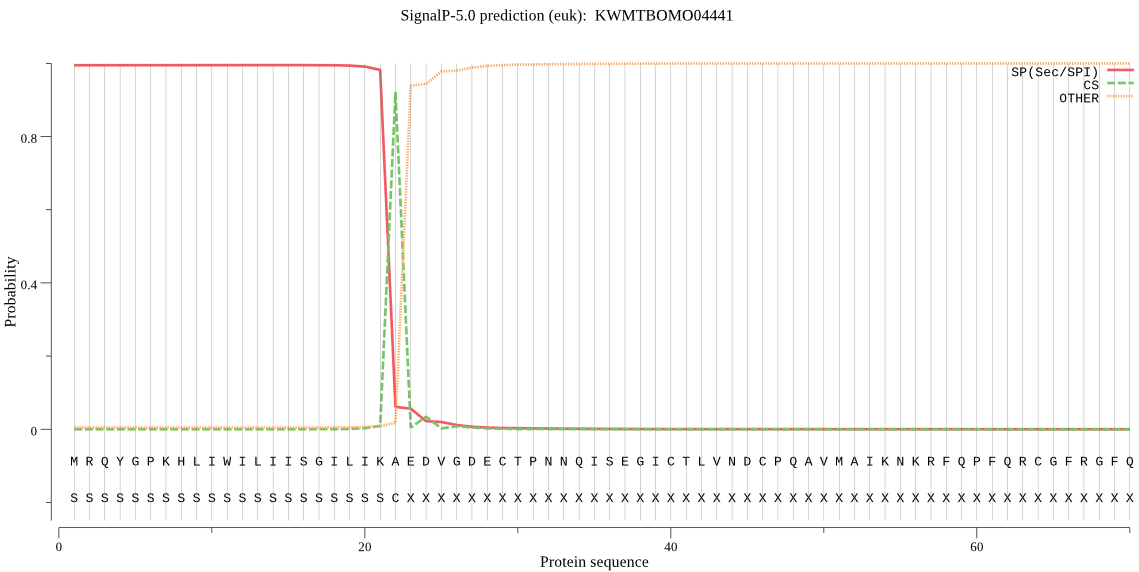
Length:
357
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0583
Exp number, first 60 AAs:
0.03947
Total prob of N-in:
0.01749
outside
1 - 357
Population Genetic Test Statistics
Pi
224.419146
Theta
188.495587
Tajima's D
0.287996
CLR
0.245532
CSRT
0.450027498625069
Interpretation
Possibly Positive selection
