Gene
KWMTBOMO04438
Pre Gene Modal
BGIBMGA005382
Annotation
DNA_cytosine-5_methyltransferase_[Bombyx_mori]
Full name
DNA (cytosine-5)-methyltransferase
+ More
Cytosine-specific methyltransferase
DNA (cytosine-5)-methyltransferase PliMCI
DNA (cytosine-5)-methyltransferase 1
Cytosine-specific methyltransferase
DNA (cytosine-5)-methyltransferase PliMCI
DNA (cytosine-5)-methyltransferase 1
Alternative Name
DNA methyltransferase PliMCI
Dnmt1
MCMT
DNA methyltransferase GgaI
Dnmt1
MCMT
DNA methyltransferase GgaI
Location in the cell
Nuclear Reliability : 3.783
Sequence
CDS
ATGCCCACTTCAACTATTACTTGCAACAGTATAACACAAGTGTGCATGGATAAGATACTTGATGGGGATGAAATAATCAAAGAAAACTCTAAGAGAAAAAGGAGCTGTGAAGAAATTGTTTCTGCCAATAATAAACGATCAAAAACAGAAGACAATGATGGACATCAAATTTCACAAGATTCTACAAGCAATGACTCTATTGAAAATCTTAACATTCCAGCAAAAAATACTAGTCTTATTGTTTCTGAAAACATCAATAATGTATATAAAAATTATGACGAGGTTGAAAATAAACCAGTATATAAAAATGGTGACCCTATCGTTGTTGAAATAGATAACAAGGATAAGCCTGGATCAGTTAACAACAATCATGACATGATTGATGATTCAGAGACCATATCTAATATTAAGAATAGCAATAATATTATCCCTGATACAGAGAAATGTAACATATGTGGCCAATTTTTAAATAATTCTGACTTAATTTATTACCAAGGTCACCCACAAGATGCTGTTGAAGAATATATTGCATTGACTAATGACAAACTTGTTTTGAGCTCTGGGGAAGATGGAGACATAATGGAACGACCCCAAACAAATATAACTGGCTTTACAATATTTGATGAGCAAGGACATTTGTGTCCAATTGATGGTGGTTTGGTAGAAAACGATGTTCGAATTTATATGTCTGGATATTTAAAGTCAATTTGCTCTGATTCATCAGAGATTGACGAAGAATCCATTCCAGTTAAAGATGTAGGTCCTATAATAGAATGGTTCATACATGGTTTCGATGGTGGAGATAGGAACTGTATAACTTTATCAACGGAATTTGGGGAATATAACTTGCTCAAACCTAGTGAAGCTTATACGCCCCTAATGAATAATTTATATGAAAAAATATGGCTTAGCAAAGTAGTCGTCGAATTCTTAGAAGAATATCACTACTTGCAACCGAGCTATGAAGATTTACTTGAAGTTGTAAGAGATTTTTCCATTCCTGAACTAAATAATAAAAAGATGACAGAAGAAATGCTACATAAACATGCCCAGTTTGTTTGTGATCAAGTAGTTAGCTTAGAAATAGAGGAAGATGATGAACCATTAATAACATTACCTTGTATGAGAGAACTTATCAAATTAATGGGCATAAAATTCGGTAAACGAAAAATACGTACCCAGATACAATACAAAAAAACAGATAAGAAAGCATGGACAAAAGCTACAACAACTCCACTTGTAAGAAAAACATTTGAAAGTTTTTTCTCAAATCAGTTAGACAAAACCAATCACGAACTGGTCTTACGAAGAAAACGATGCGGGGTTTGTGAAGCCTGTCAATTGCCTGATTGCGGCGAATGTAATGCATGTCGTGCCATGGCTAAATTCGGTGGTCATGGAAGAACTAAGAAAGCATGCGTCCGTAGATTATGTCCTAATATGGCAGTAGAACAGGCAGAAGATTCTGATCCCGATGATGAAGATGAGTATCAGCAAATATCTGAGAAAAAACAAGACAAAATAGATGATGCAGTACCAGTCAAGCTCACTGGGTCAAATAGTAAAAATTTAAAATGGATTGGTGAGCCTGTTAAAGCTGATGCCACAAAAATATATTACGAAAAAGTTGAAATTGATGGTGCTGAGCTGTGTAATGGTGACTTTGTCATGATTGAAACTTCCCAAACAAACATTCCTACACTGGTCGCTAAAGTTGTATACATGTGGAAAGAAATCCATAACCCGAAATCAGGCTATTTTCACGGAGAAGTCTTCATACGAGCCTCGGACACAGTTTTAGGAGAAGTGAGTGACCCAAGAGAAGTATTTTTGGGCGACAGATGTTGCCATGGTGCTCCGCTTTCTTCCATATTAAGAAAAGCGAATATAGAAAGGAAAGAGACGTCAGCAGATTGGTTTAAACTTGGCGGGAAAGAAGTAGACGACGAACATTTCGAGGATGATGGAAGAACTTATTTTTATAGCAAGTATTACGACAGGTTCACGTCGCGTTTTGAAGATTTACCTCCAGATCCAGCGTGTCCAAATGCTTTAAGAAAGCACAGATTCTGCCCATCTTGTGAACGTAAAACAAAACGAGATGCAAGAAACATTCCTAAAGTTTTTGAAAAATTGATTGTGAAATCCGAAATTGTTTCAGAACAAAACAGGTCGGAATGGTCATATGTAAAATGGCAAGACTTTGACTATAAGAAGGGTTGTGGTGTTTTCCTAAAACCGGGAACCTTTAAACTAAAAAATAGTATGACAAAAGCGAACACTGTTGCCAAACCGCGGTTTGAAAAAGTCGATGAAACTATTTATCCTGAATATTATAGGAAGAACGATTCTAATACGCGTGGGTCTAATATTGATACCGGGGAACCTTTTTGTGTGGGTTACATAGCTGCCGTCACTGCCGCGAGTGAGGGACCTCTAGTTGTACCGCAAGATATTTACCTGAAAGTGAATGTGCTGCTTAGACCTGAAAACACGAGCAGTAAATTTCCGCAGCACGAGGACACTAATGTTTTATATTGGACAACTGAAATAAGGGAGATTCCATTTTCGACCGTCGTTGGGCATTGTCATTTAATTTATGAACAAAATGTTCCCCAAAATATTTCACTTCAAGAATGGTTAGGAAATGATCCGTGCAGATTCTACTTCAGAATGGCGTATTGTAAGTCCACGGGAGAGTTCACGGATCTCCCGCAGAATGCCATCAGTGTTGGAAGAACAGATCGTACTAAAGATAAAGGGAAGGGGAAGGGAAAATCTACAAAAACTATTGAAACGGTTCCCGCTAAAGTTGTAGAGGAGAAAATTAGACCGTTGCGAACTTTAGACGTGTTTGCCGGTTGTGGGGGCCTATCAGAAGGCTTGCATCAAGCTGGAGTTGCAGAATGCAAGTGGGCTATTGAAAACGTGGAAGCTGCTTCTCATGCTTATTCATTAAATAATAAAAGTTGTATTGTATTTAATGAAGACTGTAATGCACTTTTGAAAACTGTAATGTCTGGTGCTAAGCACAGCGCGAATGGACTGCGGCTCCCCATGCAAGGAGAAGTGGAGCTTTTATGTGGCGGGCCACCATGTCAAGGCTTCTCCGGAATGAATCGTTTTAATTCAAGAGAGTATTCAAACTTCAAAAACTCATTAGTTGCATCGTATTTATCGTTTTGTGATTATTACAGACCTAAATATTTTATACTGGAAAACGTTCGTAACTTTGTGGCCTTTAAAAAAGGTATGGTTTTGAAACTCACTCTGCGAGCTTTGTTGGACATGGGATACCAGTGTACCTTTGGTATCTTACAAGCGGGTAACTACGGAGTTCCCCAAACTAGACGAAGGTTGATCATATTGGCGGCAGCGCCGGGTTACAATCTTCCGTTCTACCCAGAACCCACGCACGTCTTCAGTAGACGTGCTTGTACATTAACAACGACGATTGACGGCAAAAGGTTCTCGACTAATATTCACTGGGACGAGTCGGCTCCAAAAAGAACGTGCACCATTCAAGACGCCATGAGTGATTTGCCTCAAATATGTAATGGTGCTAATAAAATAGAAATCGAATATGGCTCCATGCCAGAAAGTCATTTCCAGAGATTAGTGCGAAGCAACGATGAGAATTCTAAACTACGAGATCACATCTGTAAAAACATGGCGCCATTGATTCAAGCTAGGATCAGCAGGATACCTACAACACCGGGCTCCGACTGGAGAGATCTACCCAACATATCGGTTACTCTCTCCGATGGCACTAAGTGCAAGGTTCTACAGTATCGCTACGACGACAAGCGCAATGGGCGGTCGAGCAGCGGCGCAGTGCGCGGCGTGTGCGCGTGCGCGAGCGGCCGCGCCTGCTCGCCGCTGGACAAGCAGGAGAACACGCTCATCCCGTGGTGTCTGCCGCACACCGGCAACAGGCACAACCACTGGGCCGGGCTCTACGGTCGGCTATCGTGGGGCGGGTACTTCAGCACGACAGTGACGGATCCCGAACCGATGGGCAAGCAAGGCCGGGTGCTCCACCCCGACCAGCACCGAGTCGTCTCCGTGCGAGAGTGCGCGCGCTCGCAGGGCTTCCCTGATACGTATCTCTTCGCAGGCTCCGTACAGGACAAACATCGACAGATCGGAAATGCAGTTCCACCACCTCTTGGCGCCGCTCTGGGTAGAGAAATAAAGAAAGCGCTTACTCTCAGCCTGACCACATCATGA
Protein
MPTSTITCNSITQVCMDKILDGDEIIKENSKRKRSCEEIVSANNKRSKTEDNDGHQISQDSTSNDSIENLNIPAKNTSLIVSENINNVYKNYDEVENKPVYKNGDPIVVEIDNKDKPGSVNNNHDMIDDSETISNIKNSNNIIPDTEKCNICGQFLNNSDLIYYQGHPQDAVEEYIALTNDKLVLSSGEDGDIMERPQTNITGFTIFDEQGHLCPIDGGLVENDVRIYMSGYLKSICSDSSEIDEESIPVKDVGPIIEWFIHGFDGGDRNCITLSTEFGEYNLLKPSEAYTPLMNNLYEKIWLSKVVVEFLEEYHYLQPSYEDLLEVVRDFSIPELNNKKMTEEMLHKHAQFVCDQVVSLEIEEDDEPLITLPCMRELIKLMGIKFGKRKIRTQIQYKKTDKKAWTKATTTPLVRKTFESFFSNQLDKTNHELVLRRKRCGVCEACQLPDCGECNACRAMAKFGGHGRTKKACVRRLCPNMAVEQAEDSDPDDEDEYQQISEKKQDKIDDAVPVKLTGSNSKNLKWIGEPVKADATKIYYEKVEIDGAELCNGDFVMIETSQTNIPTLVAKVVYMWKEIHNPKSGYFHGEVFIRASDTVLGEVSDPREVFLGDRCCHGAPLSSILRKANIERKETSADWFKLGGKEVDDEHFEDDGRTYFYSKYYDRFTSRFEDLPPDPACPNALRKHRFCPSCERKTKRDARNIPKVFEKLIVKSEIVSEQNRSEWSYVKWQDFDYKKGCGVFLKPGTFKLKNSMTKANTVAKPRFEKVDETIYPEYYRKNDSNTRGSNIDTGEPFCVGYIAAVTAASEGPLVVPQDIYLKVNVLLRPENTSSKFPQHEDTNVLYWTTEIREIPFSTVVGHCHLIYEQNVPQNISLQEWLGNDPCRFYFRMAYCKSTGEFTDLPQNAISVGRTDRTKDKGKGKGKSTKTIETVPAKVVEEKIRPLRTLDVFAGCGGLSEGLHQAGVAECKWAIENVEAASHAYSLNNKSCIVFNEDCNALLKTVMSGAKHSANGLRLPMQGEVELLCGGPPCQGFSGMNRFNSREYSNFKNSLVASYLSFCDYYRPKYFILENVRNFVAFKKGMVLKLTLRALLDMGYQCTFGILQAGNYGVPQTRRRLIILAAAPGYNLPFYPEPTHVFSRRACTLTTTIDGKRFSTNIHWDESAPKRTCTIQDAMSDLPQICNGANKIEIEYGSMPESHFQRLVRSNDENSKLRDHICKNMAPLIQARISRIPTTPGSDWRDLPNISVTLSDGTKCKVLQYRYDDKRNGRSSSGAVRGVCACASGRACSPLDKQENTLIPWCLPHTGNRHNHWAGLYGRLSWGGYFSTTVTDPEPMGKQGRVLHPDQHRVVSVRECARSQGFPDTYLFAGSVQDKHRQIGNAVPPPLGAALGREIKKALTLSLTTS
Summary
Description
Methylates CpG residues. Preferentially methylates hemimethylated DNA. It is responsible for maintaining methylation patterns established in development. Mediates transcriptional repression by direct binding to HDAC2. Plays a role in promoter hypermethylation and transcriptional silencing of tumor suppressor genes (TSGs) or other tumor-related genes. Also required to maintain a transcriptionally repressive state of genes in undifferentiated embryonic stem cells (ESCs). Associates at promoter regions of tumor suppressor genes (TSGs) leading to their gene silencing.
Methylates CpG residues. Preferentially methylates hemimethylated DNA. Associates with DNA replication sites in S phase maintaining the methylation pattern in the newly synthesized strand, that is essential for epigenetic inheritance. Associates with chromatin during G2 and M phases to maintain DNA methylation independently of replication. It is responsible for maintaining methylation patterns established in development. DNA methylation is coordinated with methylation of histones. Mediates transcriptional repression by direct binding to HDAC2. In association with DNMT3B and via the recruitment of CTCFL/BORIS, involved in activation of BAG1 gene expression by modulating dimethylation of promoter histone H3 at H3K4 and H3K9. Probably forms a corepressor complex required for activated KRAS-mediated promoter hypermethylation and transcriptional silencing of tumor suppressor genes (TSGs) or other tumor-related genes in colorectal cancer (CRC) cells. Also required to maintain a transcriptionally repressive state of genes in undifferentiated embryonic stem cells (ESCs). Associates at promoter regions of tumor suppressor genes (TSGs) leading to their gene silencing. Promotes tumor growth.
Methylates CpG residues. Preferentially methylates hemimethylated DNA. Associates with DNA replication sites in S phase maintaining the methylation pattern in the newly synthesized strand, that is essential for epigenetic inheritance. Associates with chromatin during G2 and M phases to maintain DNA methylation independently of replication. It is responsible for maintaining methylation patterns established in development. DNA methylation is coordinated with methylation of histones. Mediates transcriptional repression by direct binding to HDAC2. In association with DNMT3B and via the recruitment of CTCFL/BORIS, involved in activation of BAG1 gene expression by modulating dimethylation of promoter histone H3 at H3K4 and H3K9. Probably forms a corepressor complex required for activated KRAS-mediated promoter hypermethylation and transcriptional silencing of tumor suppressor genes (TSGs) or other tumor-related genes in colorectal cancer (CRC) cells. Also required to maintain a transcriptionally repressive state of genes in undifferentiated embryonic stem cells (ESCs). Associates at promoter regions of tumor suppressor genes (TSGs) leading to their gene silencing. Promotes tumor growth.
Catalytic Activity
a 2'-deoxycytidine in DNA + S-adenosyl-L-methionine = a 5-methyl-2'-deoxycytidine in DNA + H(+) + S-adenosyl-L-homocysteine
Subunit
Homodimer (By similarity). Interacts with PCNA (PubMed:9302295).
Homodimer. Forms a stable complex with E2F1, BB1 and HDAC1. Forms a complex with DMAP1 and HDAC2, with direct interaction. Interacts with the PRC2/EED-EZH2 complex. Probably part of a corepressor complex containing ZNF304, TRIM28, SETDB1 and DNMT1. Interacts with UHRF1; promoting its recruitment to hemimethylated DNA. Interacts with USP7, promoting its deubiquitination. Interacts with BAZ2A/TIP5. Interacts with PCNA. Interacts with MBD2 and MBD3. Interacts with DNMT3A and DNMT3B. Interacts with UBC9. Interacts with HDAC1. Interacts with CSNK1D.
Homodimer. Forms a stable complex with E2F1, BB1 and HDAC1. Forms a complex with DMAP1 and HDAC2, with direct interaction. Interacts with the PRC2/EED-EZH2 complex. Probably part of a corepressor complex containing ZNF304, TRIM28, SETDB1 and DNMT1. Interacts with UHRF1; promoting its recruitment to hemimethylated DNA. Interacts with USP7, promoting its deubiquitination. Interacts with BAZ2A/TIP5. Interacts with PCNA. Interacts with MBD2 and MBD3. Interacts with DNMT3A and DNMT3B. Interacts with UBC9. Interacts with HDAC1. Interacts with CSNK1D.
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. C5-methyltransferase family.
Keywords
DNA-binding
Metal-binding
Methyltransferase
Nucleus
Repeat
S-adenosyl-L-methionine
Transferase
Zinc
Zinc-finger
Complete proteome
Phosphoprotein
Reference proteome
Repressor
Transcription
Transcription regulation
Acetylation
Activator
Chromatin regulator
Isopeptide bond
Methylation
Ubl conjugation
Feature
chain DNA (cytosine-5)-methyltransferase PliMCI
Uniprot
Q5W7N6
A0A0A1GXX8
H9J790
A0A2H1WNZ3
A0A0L7L4X6
A0A2S1TXU8
+ More
A0A212FFU2 A0A194RD86 A0A0L7L3U4 A0A154PR63 A0A154PCJ8 A0A0L7RI38 A0A348ATE2 A0A2A3E789 A0A088A7M4 A0A026WZU0 K7IRF2 A0A1B6C2M8 E2C6Q5 A0A232ER49 A0A067ZXT3 A0A088AAI0 A0A221HL23 A0A2A3E1W4 A0A1B6K8F8 A0A2J7QNJ8 A0A0M9A8A2 A0A3M6TBL6 A0A195ED15 F4WNH8 A0A2B4SZP1 A0A158NTU7 A0A195B889 A0A195FXM2 A0A310SLS1 A0A023F2S5 A0A0L8GEZ1 A0A151WVE7 A0A193DUV7 A0A224XCY1 N0C9D2 J9VCT6 A0A3P9I834 A0A3P9I847 A0A3P9K564 A0A3B3HQS6 H2LLV3 C3YZQ2 A0A3P9K3W6 A0A3B4E434 W4Z639 A0A3B4CJF5 A0A1S3IR91 A0A3Q0RKD2 A0A3B3B4M6 A0A3Q0RFD3 A0A3B3B3B8 Q27746 A0A3B4ZG72 Q8IAJ7 A0A151PGK4 A0A3B4XQH7 A0A3Q0GJ98 A0A1U7S0P2 A0A1Y1N290 A0A1D5PU01 A0A3Q0GK28 Q92072 W5UBR2 A0A3Q3M6K6 A0A3P8SRD4 A0A2P8YGW3 A0A3Q3M6G6 A0A3Q1C2J2 A0A3Q1ALP3 F6QE78 A0A3P8SRQ1 A0A3Q1FFF6 A0A3Q1G8P5 A0A1V4JAG4 A0A287A8G0 A0A3P8SRI8 A0A3B4G960 W5KY99 A0A3Q1C2H1 W5KY98 A0A3Q1F9R6 A0A3P9CTE5 A0A287B7V7 A0A2Y9M0U3 A0A2Y9M696 A0A3P8QG56 A0A2Y9TG09 A0A2Y9TJE7 A0A3B3X3M6 A0A3Q2X3E7 A0A087XKB9 A0A3B3UUF0 A0A2Y9THV2 Q24K09
A0A212FFU2 A0A194RD86 A0A0L7L3U4 A0A154PR63 A0A154PCJ8 A0A0L7RI38 A0A348ATE2 A0A2A3E789 A0A088A7M4 A0A026WZU0 K7IRF2 A0A1B6C2M8 E2C6Q5 A0A232ER49 A0A067ZXT3 A0A088AAI0 A0A221HL23 A0A2A3E1W4 A0A1B6K8F8 A0A2J7QNJ8 A0A0M9A8A2 A0A3M6TBL6 A0A195ED15 F4WNH8 A0A2B4SZP1 A0A158NTU7 A0A195B889 A0A195FXM2 A0A310SLS1 A0A023F2S5 A0A0L8GEZ1 A0A151WVE7 A0A193DUV7 A0A224XCY1 N0C9D2 J9VCT6 A0A3P9I834 A0A3P9I847 A0A3P9K564 A0A3B3HQS6 H2LLV3 C3YZQ2 A0A3P9K3W6 A0A3B4E434 W4Z639 A0A3B4CJF5 A0A1S3IR91 A0A3Q0RKD2 A0A3B3B4M6 A0A3Q0RFD3 A0A3B3B3B8 Q27746 A0A3B4ZG72 Q8IAJ7 A0A151PGK4 A0A3B4XQH7 A0A3Q0GJ98 A0A1U7S0P2 A0A1Y1N290 A0A1D5PU01 A0A3Q0GK28 Q92072 W5UBR2 A0A3Q3M6K6 A0A3P8SRD4 A0A2P8YGW3 A0A3Q3M6G6 A0A3Q1C2J2 A0A3Q1ALP3 F6QE78 A0A3P8SRQ1 A0A3Q1FFF6 A0A3Q1G8P5 A0A1V4JAG4 A0A287A8G0 A0A3P8SRI8 A0A3B4G960 W5KY99 A0A3Q1C2H1 W5KY98 A0A3Q1F9R6 A0A3P9CTE5 A0A287B7V7 A0A2Y9M0U3 A0A2Y9M696 A0A3P8QG56 A0A2Y9TG09 A0A2Y9TJE7 A0A3B3X3M6 A0A3Q2X3E7 A0A087XKB9 A0A3B3UUF0 A0A2Y9THV2 Q24K09
EC Number
2.1.1.37
Pubmed
EMBL
AB194008
BAD67189.1
LC010238
BAP86925.1
BABH01013633
ODYU01010002
+ More
SOQ54793.1 JTDY01002883 KOB70543.1 MF347704 AWI66421.1 AGBW02008764 OWR52605.1 KQ460397 KPJ15240.1 JTDY01003135 KOB70090.1 KQ435012 KZC13768.1 KQ434870 KZC09572.1 KQ414585 KOC70520.1 FX985462 BBC27273.1 KZ288347 PBC27578.1 KK107061 EZA61363.1 GEDC01029530 JAS07768.1 GL453161 EFN76367.1 NNAY01002670 OXU20811.1 KF294265 AHZ08393.1 GU907553 ASM42030.1 KZ288444 PBC25685.1 GEBQ01032237 JAT07740.1 NEVH01013194 PNF30164.1 KQ435727 KOX77963.1 RCHS01003956 RMX38664.1 KQ979074 KYN22712.1 GL888237 EGI64171.1 LSMT01000002 PFX34776.1 ADTU01002818 KQ976565 KYM80462.1 KQ981215 KYN44594.1 KQ761467 OAD57514.1 GBBI01003378 JAC15334.1 KQ422062 KOF75587.1 KQ982706 KYQ51853.1 KX064246 ANN46487.1 GFTR01008788 JAW07638.1 KC210883 AGK72242.1 JX027277 AFR91943.1 GG666566 EEN54323.1 AAGJ04051615 AAGJ04051616 AAGJ04051617 Z50183 AJ495767 AJ495845 AJ495846 AJ495847 AJ495848 AJ495849 AJ495850 AJ495851 AJ495852 AJ495853 AJ495854 AJ495855 AJ496187 CAD42182.3 AKHW03000333 KYO47915.1 GEZM01014671 JAV92043.1 AADN05000499 D43920 JT410353 AHH39024.1 PYGN01000608 PSN43414.1 AAMC01032281 LSYS01008266 OPJ69161.1 AEMK02000012 AYCK01008264 AY244709 AY173048 BC114063
SOQ54793.1 JTDY01002883 KOB70543.1 MF347704 AWI66421.1 AGBW02008764 OWR52605.1 KQ460397 KPJ15240.1 JTDY01003135 KOB70090.1 KQ435012 KZC13768.1 KQ434870 KZC09572.1 KQ414585 KOC70520.1 FX985462 BBC27273.1 KZ288347 PBC27578.1 KK107061 EZA61363.1 GEDC01029530 JAS07768.1 GL453161 EFN76367.1 NNAY01002670 OXU20811.1 KF294265 AHZ08393.1 GU907553 ASM42030.1 KZ288444 PBC25685.1 GEBQ01032237 JAT07740.1 NEVH01013194 PNF30164.1 KQ435727 KOX77963.1 RCHS01003956 RMX38664.1 KQ979074 KYN22712.1 GL888237 EGI64171.1 LSMT01000002 PFX34776.1 ADTU01002818 KQ976565 KYM80462.1 KQ981215 KYN44594.1 KQ761467 OAD57514.1 GBBI01003378 JAC15334.1 KQ422062 KOF75587.1 KQ982706 KYQ51853.1 KX064246 ANN46487.1 GFTR01008788 JAW07638.1 KC210883 AGK72242.1 JX027277 AFR91943.1 GG666566 EEN54323.1 AAGJ04051615 AAGJ04051616 AAGJ04051617 Z50183 AJ495767 AJ495845 AJ495846 AJ495847 AJ495848 AJ495849 AJ495850 AJ495851 AJ495852 AJ495853 AJ495854 AJ495855 AJ496187 CAD42182.3 AKHW03000333 KYO47915.1 GEZM01014671 JAV92043.1 AADN05000499 D43920 JT410353 AHH39024.1 PYGN01000608 PSN43414.1 AAMC01032281 LSYS01008266 OPJ69161.1 AEMK02000012 AYCK01008264 AY244709 AY173048 BC114063
Proteomes
UP000005204
UP000037510
UP000007151
UP000053240
UP000076502
UP000053825
+ More
UP000242457 UP000005203 UP000053097 UP000002358 UP000008237 UP000215335 UP000235965 UP000053105 UP000275408 UP000078492 UP000007755 UP000225706 UP000005205 UP000078540 UP000078541 UP000053454 UP000075809 UP000265200 UP000265180 UP000001038 UP000001554 UP000261440 UP000007110 UP000085678 UP000261340 UP000261560 UP000261400 UP000050525 UP000261360 UP000189705 UP000000539 UP000221080 UP000261640 UP000265080 UP000245037 UP000257160 UP000008143 UP000257200 UP000190648 UP000008227 UP000261460 UP000018467 UP000265160 UP000248483 UP000265100 UP000248484 UP000261480 UP000264840 UP000028760 UP000261500 UP000009136
UP000242457 UP000005203 UP000053097 UP000002358 UP000008237 UP000215335 UP000235965 UP000053105 UP000275408 UP000078492 UP000007755 UP000225706 UP000005205 UP000078540 UP000078541 UP000053454 UP000075809 UP000265200 UP000265180 UP000001038 UP000001554 UP000261440 UP000007110 UP000085678 UP000261340 UP000261560 UP000261400 UP000050525 UP000261360 UP000189705 UP000000539 UP000221080 UP000261640 UP000265080 UP000245037 UP000257160 UP000008143 UP000257200 UP000190648 UP000008227 UP000261460 UP000018467 UP000265160 UP000248483 UP000265100 UP000248484 UP000261480 UP000264840 UP000028760 UP000261500 UP000009136
Pfam
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
Q5W7N6
A0A0A1GXX8
H9J790
A0A2H1WNZ3
A0A0L7L4X6
A0A2S1TXU8
+ More
A0A212FFU2 A0A194RD86 A0A0L7L3U4 A0A154PR63 A0A154PCJ8 A0A0L7RI38 A0A348ATE2 A0A2A3E789 A0A088A7M4 A0A026WZU0 K7IRF2 A0A1B6C2M8 E2C6Q5 A0A232ER49 A0A067ZXT3 A0A088AAI0 A0A221HL23 A0A2A3E1W4 A0A1B6K8F8 A0A2J7QNJ8 A0A0M9A8A2 A0A3M6TBL6 A0A195ED15 F4WNH8 A0A2B4SZP1 A0A158NTU7 A0A195B889 A0A195FXM2 A0A310SLS1 A0A023F2S5 A0A0L8GEZ1 A0A151WVE7 A0A193DUV7 A0A224XCY1 N0C9D2 J9VCT6 A0A3P9I834 A0A3P9I847 A0A3P9K564 A0A3B3HQS6 H2LLV3 C3YZQ2 A0A3P9K3W6 A0A3B4E434 W4Z639 A0A3B4CJF5 A0A1S3IR91 A0A3Q0RKD2 A0A3B3B4M6 A0A3Q0RFD3 A0A3B3B3B8 Q27746 A0A3B4ZG72 Q8IAJ7 A0A151PGK4 A0A3B4XQH7 A0A3Q0GJ98 A0A1U7S0P2 A0A1Y1N290 A0A1D5PU01 A0A3Q0GK28 Q92072 W5UBR2 A0A3Q3M6K6 A0A3P8SRD4 A0A2P8YGW3 A0A3Q3M6G6 A0A3Q1C2J2 A0A3Q1ALP3 F6QE78 A0A3P8SRQ1 A0A3Q1FFF6 A0A3Q1G8P5 A0A1V4JAG4 A0A287A8G0 A0A3P8SRI8 A0A3B4G960 W5KY99 A0A3Q1C2H1 W5KY98 A0A3Q1F9R6 A0A3P9CTE5 A0A287B7V7 A0A2Y9M0U3 A0A2Y9M696 A0A3P8QG56 A0A2Y9TG09 A0A2Y9TJE7 A0A3B3X3M6 A0A3Q2X3E7 A0A087XKB9 A0A3B3UUF0 A0A2Y9THV2 Q24K09
A0A212FFU2 A0A194RD86 A0A0L7L3U4 A0A154PR63 A0A154PCJ8 A0A0L7RI38 A0A348ATE2 A0A2A3E789 A0A088A7M4 A0A026WZU0 K7IRF2 A0A1B6C2M8 E2C6Q5 A0A232ER49 A0A067ZXT3 A0A088AAI0 A0A221HL23 A0A2A3E1W4 A0A1B6K8F8 A0A2J7QNJ8 A0A0M9A8A2 A0A3M6TBL6 A0A195ED15 F4WNH8 A0A2B4SZP1 A0A158NTU7 A0A195B889 A0A195FXM2 A0A310SLS1 A0A023F2S5 A0A0L8GEZ1 A0A151WVE7 A0A193DUV7 A0A224XCY1 N0C9D2 J9VCT6 A0A3P9I834 A0A3P9I847 A0A3P9K564 A0A3B3HQS6 H2LLV3 C3YZQ2 A0A3P9K3W6 A0A3B4E434 W4Z639 A0A3B4CJF5 A0A1S3IR91 A0A3Q0RKD2 A0A3B3B4M6 A0A3Q0RFD3 A0A3B3B3B8 Q27746 A0A3B4ZG72 Q8IAJ7 A0A151PGK4 A0A3B4XQH7 A0A3Q0GJ98 A0A1U7S0P2 A0A1Y1N290 A0A1D5PU01 A0A3Q0GK28 Q92072 W5UBR2 A0A3Q3M6K6 A0A3P8SRD4 A0A2P8YGW3 A0A3Q3M6G6 A0A3Q1C2J2 A0A3Q1ALP3 F6QE78 A0A3P8SRQ1 A0A3Q1FFF6 A0A3Q1G8P5 A0A1V4JAG4 A0A287A8G0 A0A3P8SRI8 A0A3B4G960 W5KY99 A0A3Q1C2H1 W5KY98 A0A3Q1F9R6 A0A3P9CTE5 A0A287B7V7 A0A2Y9M0U3 A0A2Y9M696 A0A3P8QG56 A0A2Y9TG09 A0A2Y9TJE7 A0A3B3X3M6 A0A3Q2X3E7 A0A087XKB9 A0A3B3UUF0 A0A2Y9THV2 Q24K09
PDB
4WXX
E-value=0,
Score=2896
Ontologies
PATHWAY
GO
GO:0003677
GO:0005634
GO:0003886
GO:0003682
GO:0008270
GO:0016021
GO:0070121
GO:0060216
GO:0048565
GO:0051567
GO:0010842
GO:0007368
GO:0035622
GO:0051216
GO:0044030
GO:0002088
GO:0031017
GO:0010424
GO:0010216
GO:0043045
GO:0016458
GO:0000122
GO:0007265
GO:1990841
GO:0005721
GO:0051571
GO:0071230
GO:1905931
GO:1904707
GO:1905460
GO:0005657
GO:0003723
GO:0051573
GO:0008327
GO:0010628
GO:0090309
GO:0006325
GO:0008168
GO:0004674
GO:0009058
GO:0016742
GO:0003824
GO:0090116
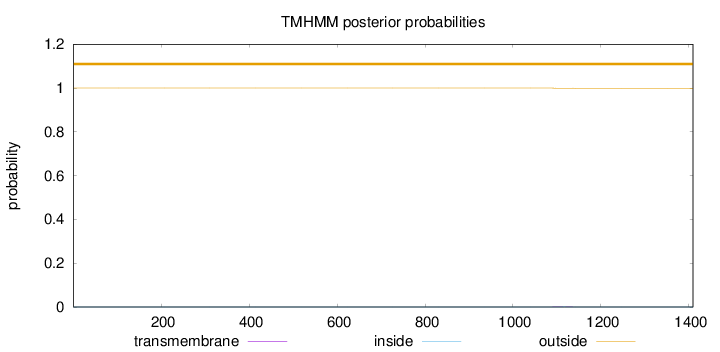
Topology
Subcellular location
Nucleus
Length:
1409
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01602
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00000
outside
1 - 1409
Population Genetic Test Statistics
Pi
182.670958
Theta
207.13532
Tajima's D
-2.346409
CLR
1.965968
CSRT
0.00264986750662467
Interpretation
Uncertain
