Pre Gene Modal
BGIBMGA005386
Annotation
PREDICTED:_arf-GAP_with_coiled-coil?_ANK_repeat_and_PH_domain-containing_protein_2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.398
Sequence
CDS
ATGCGTAACGATACAAAAATATTAGACAAAGAGATGGAGAATCGTCATACGATTGTAAACAGTAAAGACACAGTGTTGCCAAGCTGTAAAACAAATTCCGAAGGCGATTCAAAAGAGGGTTTGTTGAGTACGCCTTGCCTCAAAAATCTGCCTAGGATGCAAGGCTACTTGTTCAAGCGGACATCGAACGCTTTCAAGACTTGGAACAGAAGATGGTTTTATCTATATGACAACAGACTTGTTTACAGAAAAAGAACGGGCGAATTGAATGTAACGGTGATGGAAGAAGACCTAAGGTTATGTACAGTGAAGCCGGTTCACGACGGCGAACGAAGATTCTGTTTCGAAGTATTGTCGCCATCTAAAAGTCACATGCTCCAAGCGGACTCCGAGGAGATGTTGAGCTGCTGGATAGCGGCGCTGCAGAGCGGCATCCGCTCGGCGATACAGCAGGGCCAGAGCCGAGAGAACCCGGAGTCGCAGCTAGTGCTCGTGCCGGACGACACGAGGCTCAGCGACACCAGACGGAACACGACCAGTGTCGCCATGAAGAAAGTTAGGATTTGGGAGCAACTTCTCAGCATTCCCGGGAACAATTATTGTTGTGATTGCGGTAGTCCGAACCCGAGGTGGGCGAGCATTAATCTCGGCATCACACTATGTATAGAATGCTCGGGAATACACCGTTCGCTGGGCGTGCACGTCAGTAAAGTCAGGTCTCTGACGCTCGATGACTGGGAACCGGAAATTATAAAGGTAATGGCCGAACTAGGGAACAAATTAGTGAATTTAATATATGAAGCTAACACAGAGGGCGCGACTGTTGCCAGAGCGACGCCGGATTGTGAAACTTGTGTTCGCGAAGCGTGGATCCGTGCTAAGTACGTCTCGCTGATGTTTGTACGAGAGCTCGTGAACTTTGATGCCCTCGAACAGTCTCCTCTCAAGTCCGGCAGACGCTGGTCCGTTAGAAGAGCTCGACGAAGGTAA
Protein
MRNDTKILDKEMENRHTIVNSKDTVLPSCKTNSEGDSKEGLLSTPCLKNLPRMQGYLFKRTSNAFKTWNRRWFYLYDNRLVYRKRTGELNVTVMEEDLRLCTVKPVHDGERRFCFEVLSPSKSHMLQADSEEMLSCWIAALQSGIRSAIQQGQSRENPESQLVLVPDDTRLSDTRRNTTSVAMKKVRIWEQLLSIPGNNYCCDCGSPNPRWASINLGITLCIECSGIHRSLGVHVSKVRSLTLDDWEPEIIKVMAELGNKLVNLIYEANTEGATVARATPDCETCVREAWIRAKYVSLMFVRELVNFDALEQSPLKSGRRWSVRRARRR
Summary
Uniprot
A0A2H1WK09
A0A3S2M6K6
A0A2A4J2N2
A0A1E1WP67
A0A194RH36
A0A194PY68
+ More
D6X007 V5G2T3 A0A1W4XCI4 A0A067QVP5 A0A0C9RAI7 A0A1Y1L0G4 U4TZ47 A0A026VW25 A0A1B6I8C3 A0A1B6JJY1 A0A0J7L2Q0 N6SY22 E2C8T5 A0A154PSJ2 E9IVA0 E0W416 A0A1B6G7X6 A0A195C0F7 A0A0L7R3V4 A0A088AQD7 A0A151XHS4 A0A195FJ28 A0A2A3EAS8 A0A151I1C4 A0A158NDC8 A0A195DIU9 A0A2J7RRG9 F4WSP8 A0A2S2QYY7 A0A1Z5L5T8 A0A1E1XLH1 A0A147BQ29 A0A0A9YW47 A0A224YNB7 A0A131YD08 A0A131YXS2 A0A224XK51 A0A023F1X2 T1HYS0 L7MLR8 A0A146LN58 A0A1Q3F8P6 A0A1Q3F8K6 A0A2S2P373 B0WGF7 A0A1L8DLQ1 A0A1Q3F8V5 A0A2P8YQI4 A0A2J7RRG6 A0A1I8MUL5 A0A1Q3F8W4 W8BCJ3 A0A182GI07 A0A0A1XLV6 A0A2S2P461 A0A1B0AA11 A0A2R7W9G8 Q17GT7 A0A2H8TL56 B4HFB4 A0A1I8NN18 A0A0K2TS28 A0A1A9V7L7 A0A034VCF6 A0A1B0FKP7 Q8T0R1 A0A0K8UT23 A0A3R7PUZ4 A0A0K8W9W7 B3P7G1 B4PN46 A0A1A9YPY6 A0A1W4UQN6 Q9VCQ6 B4R1D5 A0A0K8UMX9 A0A0K8V1I0 A0A1I8MUK8 A0A0K8UW58 W8AQ27 A0A0K8VBN8 A0A0K8VGF7 A0A3B0JLI3 A0A0P6JSK4 B3LV47 A0A1S4F367 A0A336N3D1 A0A210QD91 A0A1I8NN22 A0A0K8VNT4 A0A034V833 A0A336MYX3 A0A0K8U504 A0A0R1E8R9
D6X007 V5G2T3 A0A1W4XCI4 A0A067QVP5 A0A0C9RAI7 A0A1Y1L0G4 U4TZ47 A0A026VW25 A0A1B6I8C3 A0A1B6JJY1 A0A0J7L2Q0 N6SY22 E2C8T5 A0A154PSJ2 E9IVA0 E0W416 A0A1B6G7X6 A0A195C0F7 A0A0L7R3V4 A0A088AQD7 A0A151XHS4 A0A195FJ28 A0A2A3EAS8 A0A151I1C4 A0A158NDC8 A0A195DIU9 A0A2J7RRG9 F4WSP8 A0A2S2QYY7 A0A1Z5L5T8 A0A1E1XLH1 A0A147BQ29 A0A0A9YW47 A0A224YNB7 A0A131YD08 A0A131YXS2 A0A224XK51 A0A023F1X2 T1HYS0 L7MLR8 A0A146LN58 A0A1Q3F8P6 A0A1Q3F8K6 A0A2S2P373 B0WGF7 A0A1L8DLQ1 A0A1Q3F8V5 A0A2P8YQI4 A0A2J7RRG6 A0A1I8MUL5 A0A1Q3F8W4 W8BCJ3 A0A182GI07 A0A0A1XLV6 A0A2S2P461 A0A1B0AA11 A0A2R7W9G8 Q17GT7 A0A2H8TL56 B4HFB4 A0A1I8NN18 A0A0K2TS28 A0A1A9V7L7 A0A034VCF6 A0A1B0FKP7 Q8T0R1 A0A0K8UT23 A0A3R7PUZ4 A0A0K8W9W7 B3P7G1 B4PN46 A0A1A9YPY6 A0A1W4UQN6 Q9VCQ6 B4R1D5 A0A0K8UMX9 A0A0K8V1I0 A0A1I8MUK8 A0A0K8UW58 W8AQ27 A0A0K8VBN8 A0A0K8VGF7 A0A3B0JLI3 A0A0P6JSK4 B3LV47 A0A1S4F367 A0A336N3D1 A0A210QD91 A0A1I8NN22 A0A0K8VNT4 A0A034V833 A0A336MYX3 A0A0K8U504 A0A0R1E8R9
Pubmed
26354079
18362917
19820115
24845553
28004739
23537049
+ More
24508170 30249741 20798317 21282665 20566863 21347285 21719571 28528879 29209593 29652888 25401762 28797301 26830274 25474469 25576852 26823975 29403074 25315136 24495485 26483478 25830018 17510324 17994087 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26999592 28812685
24508170 30249741 20798317 21282665 20566863 21347285 21719571 28528879 29209593 29652888 25401762 28797301 26830274 25474469 25576852 26823975 29403074 25315136 24495485 26483478 25830018 17510324 17994087 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26999592 28812685
EMBL
ODYU01009153
SOQ53378.1
RSAL01000025
RVE52141.1
NWSH01003543
PCG66129.1
+ More
GDQN01002235 JAT88819.1 KQ460397 KPJ15246.1 KQ459589 KPI97694.1 KQ971372 EFA10505.1 GALX01004141 JAB64325.1 KK852890 KDR14272.1 GBYB01013474 JAG83241.1 GEZM01068131 JAV67159.1 KB631874 ERL86864.1 KK107737 QOIP01000013 EZA47962.1 RLU15490.1 GECU01024547 JAS83159.1 GECU01008110 JAS99596.1 LBMM01001089 KMQ96943.1 APGK01052697 KB741213 ENN72674.1 GL453694 EFN75652.1 KQ435036 KZC14080.1 GL766194 EFZ15490.1 DS235886 EEB20372.1 GECZ01011213 JAS58556.1 KQ978379 KYM94389.1 KQ414661 KOC65557.1 KQ982130 KYQ59838.1 KQ981560 KYN39989.1 KZ288317 PBC28296.1 KQ976599 KYM79511.1 ADTU01012601 KQ980800 KYN12813.1 NEVH01000606 PNF43424.1 GL888326 EGI62791.1 GGMS01013763 MBY82966.1 GFJQ02004330 JAW02640.1 GFAA01003234 JAU00201.1 GEGO01002528 JAR92876.1 GBHO01009824 GBHO01009823 JAG33780.1 JAG33781.1 GFPF01006093 MAA17239.1 GEDV01012207 JAP76350.1 GEDV01005282 JAP83275.1 GFTR01007903 JAW08523.1 GBBI01003170 JAC15542.1 ACPB03018499 GACK01000801 JAA64233.1 GDHC01010407 JAQ08222.1 GFDL01011123 JAV23922.1 GFDL01011144 JAV23901.1 GGMR01010717 MBY23336.1 DS231926 EDS26954.1 GFDF01006691 JAV07393.1 GFDL01011040 JAV24005.1 PYGN01000431 PSN46505.1 PNF43422.1 GFDL01011078 JAV23967.1 GAMC01015664 JAB90891.1 JXUM01011936 JXUM01011937 JXUM01011938 JXUM01011939 JXUM01011940 JXUM01011941 JXUM01011942 KQ560341 KXJ82961.1 GBXI01002719 JAD11573.1 GGMR01011591 MBY24210.1 KK854417 PTY15690.1 CH477256 EAT45850.1 GFXV01002597 MBW14402.1 CH480815 EDW43292.1 HACA01011462 CDW28823.1 GAKP01019477 JAC39475.1 CCAG010000730 AY069114 AAL39259.1 GDHF01022646 JAI29668.1 QCYY01000697 ROT83368.1 GDHF01004331 JAI47983.1 CH954182 EDV54050.1 CM000160 EDW98101.1 AE014297 AF254071 BT122085 AAF56100.1 AAF64529.1 ADE06691.1 CM000364 EDX14021.1 GDHF01024378 JAI27936.1 GDHF01019560 JAI32754.1 GDHF01021503 JAI30811.1 GAMC01015665 JAB90890.1 GDHF01016072 JAI36242.1 GDHF01014348 JAI37966.1 OUUW01000005 SPP81192.1 GDUN01000349 JAN95570.1 CH902617 EDV42519.1 UFQT01004194 SSX35563.1 NEDP02004099 OWF46700.1 GDHF01011772 JAI40542.1 GAKP01019476 JAC39476.1 SSX35562.1 GDHF01030651 JAI21663.1 KRK04113.1
GDQN01002235 JAT88819.1 KQ460397 KPJ15246.1 KQ459589 KPI97694.1 KQ971372 EFA10505.1 GALX01004141 JAB64325.1 KK852890 KDR14272.1 GBYB01013474 JAG83241.1 GEZM01068131 JAV67159.1 KB631874 ERL86864.1 KK107737 QOIP01000013 EZA47962.1 RLU15490.1 GECU01024547 JAS83159.1 GECU01008110 JAS99596.1 LBMM01001089 KMQ96943.1 APGK01052697 KB741213 ENN72674.1 GL453694 EFN75652.1 KQ435036 KZC14080.1 GL766194 EFZ15490.1 DS235886 EEB20372.1 GECZ01011213 JAS58556.1 KQ978379 KYM94389.1 KQ414661 KOC65557.1 KQ982130 KYQ59838.1 KQ981560 KYN39989.1 KZ288317 PBC28296.1 KQ976599 KYM79511.1 ADTU01012601 KQ980800 KYN12813.1 NEVH01000606 PNF43424.1 GL888326 EGI62791.1 GGMS01013763 MBY82966.1 GFJQ02004330 JAW02640.1 GFAA01003234 JAU00201.1 GEGO01002528 JAR92876.1 GBHO01009824 GBHO01009823 JAG33780.1 JAG33781.1 GFPF01006093 MAA17239.1 GEDV01012207 JAP76350.1 GEDV01005282 JAP83275.1 GFTR01007903 JAW08523.1 GBBI01003170 JAC15542.1 ACPB03018499 GACK01000801 JAA64233.1 GDHC01010407 JAQ08222.1 GFDL01011123 JAV23922.1 GFDL01011144 JAV23901.1 GGMR01010717 MBY23336.1 DS231926 EDS26954.1 GFDF01006691 JAV07393.1 GFDL01011040 JAV24005.1 PYGN01000431 PSN46505.1 PNF43422.1 GFDL01011078 JAV23967.1 GAMC01015664 JAB90891.1 JXUM01011936 JXUM01011937 JXUM01011938 JXUM01011939 JXUM01011940 JXUM01011941 JXUM01011942 KQ560341 KXJ82961.1 GBXI01002719 JAD11573.1 GGMR01011591 MBY24210.1 KK854417 PTY15690.1 CH477256 EAT45850.1 GFXV01002597 MBW14402.1 CH480815 EDW43292.1 HACA01011462 CDW28823.1 GAKP01019477 JAC39475.1 CCAG010000730 AY069114 AAL39259.1 GDHF01022646 JAI29668.1 QCYY01000697 ROT83368.1 GDHF01004331 JAI47983.1 CH954182 EDV54050.1 CM000160 EDW98101.1 AE014297 AF254071 BT122085 AAF56100.1 AAF64529.1 ADE06691.1 CM000364 EDX14021.1 GDHF01024378 JAI27936.1 GDHF01019560 JAI32754.1 GDHF01021503 JAI30811.1 GAMC01015665 JAB90890.1 GDHF01016072 JAI36242.1 GDHF01014348 JAI37966.1 OUUW01000005 SPP81192.1 GDUN01000349 JAN95570.1 CH902617 EDV42519.1 UFQT01004194 SSX35563.1 NEDP02004099 OWF46700.1 GDHF01011772 JAI40542.1 GAKP01019476 JAC39476.1 SSX35562.1 GDHF01030651 JAI21663.1 KRK04113.1
Proteomes
UP000283053
UP000218220
UP000053240
UP000053268
UP000007266
UP000192223
+ More
UP000027135 UP000030742 UP000053097 UP000279307 UP000036403 UP000019118 UP000008237 UP000076502 UP000009046 UP000078542 UP000053825 UP000005203 UP000075809 UP000078541 UP000242457 UP000078540 UP000005205 UP000078492 UP000235965 UP000007755 UP000015103 UP000002320 UP000245037 UP000095301 UP000069940 UP000249989 UP000092445 UP000008820 UP000001292 UP000095300 UP000078200 UP000092444 UP000283509 UP000008711 UP000002282 UP000092443 UP000192221 UP000000803 UP000000304 UP000268350 UP000007801 UP000242188
UP000027135 UP000030742 UP000053097 UP000279307 UP000036403 UP000019118 UP000008237 UP000076502 UP000009046 UP000078542 UP000053825 UP000005203 UP000075809 UP000078541 UP000242457 UP000078540 UP000005205 UP000078492 UP000235965 UP000007755 UP000015103 UP000002320 UP000245037 UP000095301 UP000069940 UP000249989 UP000092445 UP000008820 UP000001292 UP000095300 UP000078200 UP000092444 UP000283509 UP000008711 UP000002282 UP000092443 UP000192221 UP000000803 UP000000304 UP000268350 UP000007801 UP000242188
PRIDE
Interpro
CDD
ProteinModelPortal
A0A2H1WK09
A0A3S2M6K6
A0A2A4J2N2
A0A1E1WP67
A0A194RH36
A0A194PY68
+ More
D6X007 V5G2T3 A0A1W4XCI4 A0A067QVP5 A0A0C9RAI7 A0A1Y1L0G4 U4TZ47 A0A026VW25 A0A1B6I8C3 A0A1B6JJY1 A0A0J7L2Q0 N6SY22 E2C8T5 A0A154PSJ2 E9IVA0 E0W416 A0A1B6G7X6 A0A195C0F7 A0A0L7R3V4 A0A088AQD7 A0A151XHS4 A0A195FJ28 A0A2A3EAS8 A0A151I1C4 A0A158NDC8 A0A195DIU9 A0A2J7RRG9 F4WSP8 A0A2S2QYY7 A0A1Z5L5T8 A0A1E1XLH1 A0A147BQ29 A0A0A9YW47 A0A224YNB7 A0A131YD08 A0A131YXS2 A0A224XK51 A0A023F1X2 T1HYS0 L7MLR8 A0A146LN58 A0A1Q3F8P6 A0A1Q3F8K6 A0A2S2P373 B0WGF7 A0A1L8DLQ1 A0A1Q3F8V5 A0A2P8YQI4 A0A2J7RRG6 A0A1I8MUL5 A0A1Q3F8W4 W8BCJ3 A0A182GI07 A0A0A1XLV6 A0A2S2P461 A0A1B0AA11 A0A2R7W9G8 Q17GT7 A0A2H8TL56 B4HFB4 A0A1I8NN18 A0A0K2TS28 A0A1A9V7L7 A0A034VCF6 A0A1B0FKP7 Q8T0R1 A0A0K8UT23 A0A3R7PUZ4 A0A0K8W9W7 B3P7G1 B4PN46 A0A1A9YPY6 A0A1W4UQN6 Q9VCQ6 B4R1D5 A0A0K8UMX9 A0A0K8V1I0 A0A1I8MUK8 A0A0K8UW58 W8AQ27 A0A0K8VBN8 A0A0K8VGF7 A0A3B0JLI3 A0A0P6JSK4 B3LV47 A0A1S4F367 A0A336N3D1 A0A210QD91 A0A1I8NN22 A0A0K8VNT4 A0A034V833 A0A336MYX3 A0A0K8U504 A0A0R1E8R9
D6X007 V5G2T3 A0A1W4XCI4 A0A067QVP5 A0A0C9RAI7 A0A1Y1L0G4 U4TZ47 A0A026VW25 A0A1B6I8C3 A0A1B6JJY1 A0A0J7L2Q0 N6SY22 E2C8T5 A0A154PSJ2 E9IVA0 E0W416 A0A1B6G7X6 A0A195C0F7 A0A0L7R3V4 A0A088AQD7 A0A151XHS4 A0A195FJ28 A0A2A3EAS8 A0A151I1C4 A0A158NDC8 A0A195DIU9 A0A2J7RRG9 F4WSP8 A0A2S2QYY7 A0A1Z5L5T8 A0A1E1XLH1 A0A147BQ29 A0A0A9YW47 A0A224YNB7 A0A131YD08 A0A131YXS2 A0A224XK51 A0A023F1X2 T1HYS0 L7MLR8 A0A146LN58 A0A1Q3F8P6 A0A1Q3F8K6 A0A2S2P373 B0WGF7 A0A1L8DLQ1 A0A1Q3F8V5 A0A2P8YQI4 A0A2J7RRG6 A0A1I8MUL5 A0A1Q3F8W4 W8BCJ3 A0A182GI07 A0A0A1XLV6 A0A2S2P461 A0A1B0AA11 A0A2R7W9G8 Q17GT7 A0A2H8TL56 B4HFB4 A0A1I8NN18 A0A0K2TS28 A0A1A9V7L7 A0A034VCF6 A0A1B0FKP7 Q8T0R1 A0A0K8UT23 A0A3R7PUZ4 A0A0K8W9W7 B3P7G1 B4PN46 A0A1A9YPY6 A0A1W4UQN6 Q9VCQ6 B4R1D5 A0A0K8UMX9 A0A0K8V1I0 A0A1I8MUK8 A0A0K8UW58 W8AQ27 A0A0K8VBN8 A0A0K8VGF7 A0A3B0JLI3 A0A0P6JSK4 B3LV47 A0A1S4F367 A0A336N3D1 A0A210QD91 A0A1I8NN22 A0A0K8VNT4 A0A034V833 A0A336MYX3 A0A0K8U504 A0A0R1E8R9
PDB
4F1P
E-value=7.57504e-39,
Score=402
Ontologies
GO
Topology
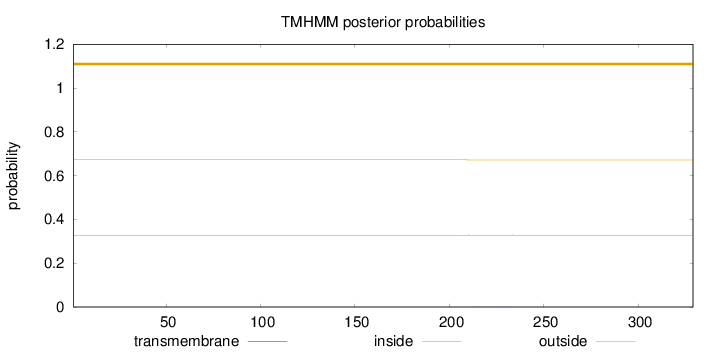
Length:
329
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00868999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.32806
outside
1 - 329
Population Genetic Test Statistics
Pi
210.233424
Theta
174.393772
Tajima's D
0.551004
CLR
0.009399
CSRT
0.526973651317434
Interpretation
Uncertain
