Gene
KWMTBOMO04431
Pre Gene Modal
BGIBMGA005388
Annotation
PREDICTED:_transmembrane_protein_19_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.7
Sequence
CDS
ATGTCCGAAAAGGAAACTAAATTCATTAAGAATGAACACGTATTAACCGTGGCTTTGTGCTCCATAGCAATACCACTGTCTATGTCTATGTGGATAATTAATATAATTTATTCCAAATATCTTGCAGAAAATTCGAGTTATGATGAACCTATTGTTATACCACCAACAAAATGGATGGTATCATGTTTTATTCCCCTTCTTATAGCAGTTTATGGATACAAGAAGAGAAGCATAAATTTAAGTGGAGCAGTTAGTGGGTTACTTATAGCATTCATTTTAACACTTAGTAGTTATGGATTTTTAGCTGCTATTTTCACATTTTTCATTACTTCATCAAAAGCTACTAAAGTAAAGTCACATTTGAAGAGGAAATTTGAAGCTGATTTTAAGGAAGGTGGACAAAGGAATTGGATACAAGTGCTGTGTAATGGTGGTATGGCAGCTCAGCTTGCATTGTTGTATCTATTGGATGTGGGATCATCCGAGCGCCCTATTGATTTTGTGAAGGATTACAGATCTTCTTGGTTGTCTATTGGAATTTTAGGTGTTTTTGCATGCTGCAATGGTGACACATGGGCTTCAGAAATTGGAACAGTTTTTTCTAGTTCAGATCCATATTTAATTACATCTTGGAAGAAAGTGCCTAGGGGCACAAATGGTGGTGTCACTTTTATTGGAACAGTATTTAGTGGTCTTGGAGGATTTGTGATTGGATTGAGCCAGTATTTTGTACTTTACTTGTTCTCAGACAAAAGTTCTTGGGATTATGCTCCACCACAATGGCCTATTATACTGATGGGCACAATTTCTGGTTTATTAGGCAGTTTAGTAGATTCTATATTGGGAGCAACTTTGCAGTTTTCAGGTTTGAACAAAGAAGGAAAAATTGTAAATCATTCCTATGACGCAGTTAAACATATCTCTGGCTGGAATATTCTAGACAATCACAGTGTAAACCTAATATCAACTATCATTATAGGCTATCTTCGGTACATCACGCTATGA
Protein
MSEKETKFIKNEHVLTVALCSIAIPLSMSMWIINIIYSKYLAENSSYDEPIVIPPTKWMVSCFIPLLIAVYGYKKRSINLSGAVSGLLIAFILTLSSYGFLAAIFTFFITSSKATKVKSHLKRKFEADFKEGGQRNWIQVLCNGGMAAQLALLYLLDVGSSERPIDFVKDYRSSWLSIGILGVFACCNGDTWASEIGTVFSSSDPYLITSWKKVPRGTNGGVTFIGTVFSGLGGFVIGLSQYFVLYLFSDKSSWDYAPPQWPIILMGTISGLLGSLVDSILGATLQFSGLNKEGKIVNHSYDAVKHISGWNILDNHSVNLISTIIIGYLRYITL
Summary
Uniprot
A0A2H1WK07
A0A194RCB9
S4NIZ4
A0A3S2PI88
A0A194PXE6
A0A212EH75
+ More
A0A0L7LD84 A0A182FUW4 A0A182MYI3 A0A2M4AUL5 J9JQ79 A0A2M4BTZ1 A0A182Y1U6 A0A182MTN9 A0A182SCX5 E2BWX7 A0A182JNY9 A0A026WSJ5 A0A182W978 A0A182PBP9 W5JCI4 E2AKI6 A0A2J7R192 A0A182USA6 Q7Q059 A0A182TWZ7 A0A182HQ12 A0A182XD82 A0A182KSP9 A0A1I8PEQ2 A0A195BHN7 A0A1B6DT90 A0A158NJW3 A0A195F414 A0A067R026 A0A084WHY4 Q2M024 B4H8Y6 A0A182JJF8 A0A3B0J955 A0A0L7QNF3 F4WIN5 A0A151XE27 A0A154PMB8 B4KZP2 A0A182R5F8 Q172X0 B0WTM0 A0A1L8D9E0 A0A088A7H5 A0A1L8D8Z9 A0A182QDF4 K7J017 A0A1S4FG33 E0W1N2 A0A0M8ZQN4 A0A1W7R551 A0A2A3EDA6 A0A1I8MKQ1 A0A0L0BVE1 A0A232EPW0 A0A1Q3F9F6 T1PIW8 T1HYQ3 B4LCH1 A0A151I852 A0A069DYL5 A0A0P4W2X8 A0A0K8TQ39 B4PHE2 A0A1B6FGK0 A0A224XNL5 W8C2H0 A0A0M4ELX5 E9IMF5 B4J177 A0A1A9WEM5 A0A023F438 A0A336MJW5 A0A1B6KHV0 A0A0K8TX67 A0A195DV96 A0A0K8VXS7 A0A3S3PIU9 Q9VU64 A0A2S2QVY9 A0A1I8PEZ9 A0A0C9Q1K6 A0A0K8W8Q5 B4HGS0 A0A1A9Y0W9 A0A1B0B6V8 A0A1A9Y0W8 A0A1B0AI87 Q8IQJ7 D3TNA2 A0A1A9V3T8 B3NHE3 Q6AWL9 L7M380
A0A0L7LD84 A0A182FUW4 A0A182MYI3 A0A2M4AUL5 J9JQ79 A0A2M4BTZ1 A0A182Y1U6 A0A182MTN9 A0A182SCX5 E2BWX7 A0A182JNY9 A0A026WSJ5 A0A182W978 A0A182PBP9 W5JCI4 E2AKI6 A0A2J7R192 A0A182USA6 Q7Q059 A0A182TWZ7 A0A182HQ12 A0A182XD82 A0A182KSP9 A0A1I8PEQ2 A0A195BHN7 A0A1B6DT90 A0A158NJW3 A0A195F414 A0A067R026 A0A084WHY4 Q2M024 B4H8Y6 A0A182JJF8 A0A3B0J955 A0A0L7QNF3 F4WIN5 A0A151XE27 A0A154PMB8 B4KZP2 A0A182R5F8 Q172X0 B0WTM0 A0A1L8D9E0 A0A088A7H5 A0A1L8D8Z9 A0A182QDF4 K7J017 A0A1S4FG33 E0W1N2 A0A0M8ZQN4 A0A1W7R551 A0A2A3EDA6 A0A1I8MKQ1 A0A0L0BVE1 A0A232EPW0 A0A1Q3F9F6 T1PIW8 T1HYQ3 B4LCH1 A0A151I852 A0A069DYL5 A0A0P4W2X8 A0A0K8TQ39 B4PHE2 A0A1B6FGK0 A0A224XNL5 W8C2H0 A0A0M4ELX5 E9IMF5 B4J177 A0A1A9WEM5 A0A023F438 A0A336MJW5 A0A1B6KHV0 A0A0K8TX67 A0A195DV96 A0A0K8VXS7 A0A3S3PIU9 Q9VU64 A0A2S2QVY9 A0A1I8PEZ9 A0A0C9Q1K6 A0A0K8W8Q5 B4HGS0 A0A1A9Y0W9 A0A1B0B6V8 A0A1A9Y0W8 A0A1B0AI87 Q8IQJ7 D3TNA2 A0A1A9V3T8 B3NHE3 Q6AWL9 L7M380
Pubmed
26354079
23622113
22118469
26227816
25244985
20798317
+ More
24508170 30249741 20920257 23761445 12364791 20966253 21347285 24845553 24438588 15632085 17994087 21719571 17510324 20075255 20566863 25315136 26108605 28648823 26334808 27129103 26369729 17550304 24495485 21282665 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 18057021 25576852
24508170 30249741 20920257 23761445 12364791 20966253 21347285 24845553 24438588 15632085 17994087 21719571 17510324 20075255 20566863 25315136 26108605 28648823 26334808 27129103 26369729 17550304 24495485 21282665 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 18057021 25576852
EMBL
ODYU01009153
SOQ53379.1
KQ460397
KPJ15247.1
GAIX01013879
JAA78681.1
+ More
RSAL01000025 RVE52143.1 KQ459589 KPI97693.1 AGBW02014942 OWR40839.1 JTDY01001577 KOB73478.1 GGFK01011149 MBW44470.1 ABLF02033194 ABLF02033200 GGFJ01007270 MBW56411.1 AXCM01002401 GL451188 EFN79841.1 KK107135 QOIP01000006 EZA58064.1 RLU21931.1 ADMH02001912 ETN60539.1 GL440281 EFN66037.1 NEVH01008208 PNF34594.1 AAAB01008986 EAA00240.3 APCN01003288 KQ976465 KYM84315.1 GEDC01011392 GEDC01008410 JAS25906.1 JAS28888.1 ADTU01018427 KQ981820 KYN35320.1 KK852804 KDR16175.1 ATLV01023896 KE525347 KFB49828.1 CH379069 EAL31112.1 CH479226 EDW35201.1 AXCP01006759 OUUW01000002 SPP76452.1 KQ414851 KOC60172.1 GL888176 EGI65971.1 KQ982254 KYQ58615.1 KQ434978 KZC13031.1 CH933809 EDW18998.1 CH477430 EAT41058.1 EJY57675.1 DS232089 EDS34473.1 GFDF01011117 JAV02967.1 GFDF01011118 JAV02966.1 AXCN02001041 DS235872 EEB19538.1 KQ435960 KOX67996.1 GEHC01001348 JAV46297.1 KZ288279 PBC29698.1 JRES01001278 KNC24007.1 NNAY01002882 OXU20390.1 GFDL01010869 JAV24176.1 KA648752 AFP63381.1 ACPB03018501 CH940647 EDW69834.1 KQ978390 KYM94290.1 GBGD01002180 JAC86709.1 GDKW01000746 JAI55849.1 GDAI01001119 JAI16484.1 CM000159 EDW94403.1 GECZ01020443 JAS49326.1 GFTR01005018 JAW11408.1 GAMC01003132 JAC03424.1 CP012525 ALC44547.1 GL764146 EFZ18246.1 CH916366 EDV97946.1 GBBI01002963 JAC15749.1 UFQT01001439 UFQT01002855 SSX30576.1 SSX34157.1 GEBQ01028968 JAT11009.1 GDHF01033262 JAI19052.1 KQ980304 KYN16771.1 GDHF01008655 JAI43659.1 NCKU01002085 RWS10469.1 AE014296 AY061341 AAF49826.1 AAL28889.1 GGMS01012713 MBY81916.1 GBYB01007763 JAG77530.1 GDHF01004900 JAI47414.1 CH480815 EDW41378.1 JXJN01009256 BT046137 AAN11833.1 ACI46525.1 CCAG010000259 EZ422904 ADD19180.1 CH954178 EDV51669.1 KQS43887.1 BT015229 AAT94458.1 GACK01007466 JAA57568.1
RSAL01000025 RVE52143.1 KQ459589 KPI97693.1 AGBW02014942 OWR40839.1 JTDY01001577 KOB73478.1 GGFK01011149 MBW44470.1 ABLF02033194 ABLF02033200 GGFJ01007270 MBW56411.1 AXCM01002401 GL451188 EFN79841.1 KK107135 QOIP01000006 EZA58064.1 RLU21931.1 ADMH02001912 ETN60539.1 GL440281 EFN66037.1 NEVH01008208 PNF34594.1 AAAB01008986 EAA00240.3 APCN01003288 KQ976465 KYM84315.1 GEDC01011392 GEDC01008410 JAS25906.1 JAS28888.1 ADTU01018427 KQ981820 KYN35320.1 KK852804 KDR16175.1 ATLV01023896 KE525347 KFB49828.1 CH379069 EAL31112.1 CH479226 EDW35201.1 AXCP01006759 OUUW01000002 SPP76452.1 KQ414851 KOC60172.1 GL888176 EGI65971.1 KQ982254 KYQ58615.1 KQ434978 KZC13031.1 CH933809 EDW18998.1 CH477430 EAT41058.1 EJY57675.1 DS232089 EDS34473.1 GFDF01011117 JAV02967.1 GFDF01011118 JAV02966.1 AXCN02001041 DS235872 EEB19538.1 KQ435960 KOX67996.1 GEHC01001348 JAV46297.1 KZ288279 PBC29698.1 JRES01001278 KNC24007.1 NNAY01002882 OXU20390.1 GFDL01010869 JAV24176.1 KA648752 AFP63381.1 ACPB03018501 CH940647 EDW69834.1 KQ978390 KYM94290.1 GBGD01002180 JAC86709.1 GDKW01000746 JAI55849.1 GDAI01001119 JAI16484.1 CM000159 EDW94403.1 GECZ01020443 JAS49326.1 GFTR01005018 JAW11408.1 GAMC01003132 JAC03424.1 CP012525 ALC44547.1 GL764146 EFZ18246.1 CH916366 EDV97946.1 GBBI01002963 JAC15749.1 UFQT01001439 UFQT01002855 SSX30576.1 SSX34157.1 GEBQ01028968 JAT11009.1 GDHF01033262 JAI19052.1 KQ980304 KYN16771.1 GDHF01008655 JAI43659.1 NCKU01002085 RWS10469.1 AE014296 AY061341 AAF49826.1 AAL28889.1 GGMS01012713 MBY81916.1 GBYB01007763 JAG77530.1 GDHF01004900 JAI47414.1 CH480815 EDW41378.1 JXJN01009256 BT046137 AAN11833.1 ACI46525.1 CCAG010000259 EZ422904 ADD19180.1 CH954178 EDV51669.1 KQS43887.1 BT015229 AAT94458.1 GACK01007466 JAA57568.1
Proteomes
UP000053240
UP000283053
UP000053268
UP000007151
UP000037510
UP000069272
+ More
UP000075884 UP000007819 UP000076408 UP000075883 UP000075901 UP000008237 UP000075881 UP000053097 UP000279307 UP000075920 UP000075885 UP000000673 UP000000311 UP000235965 UP000075903 UP000007062 UP000075902 UP000075840 UP000076407 UP000075882 UP000095300 UP000078540 UP000005205 UP000078541 UP000027135 UP000030765 UP000001819 UP000008744 UP000075880 UP000268350 UP000053825 UP000007755 UP000075809 UP000076502 UP000009192 UP000075900 UP000008820 UP000002320 UP000005203 UP000075886 UP000002358 UP000009046 UP000053105 UP000242457 UP000095301 UP000037069 UP000215335 UP000015103 UP000008792 UP000078542 UP000002282 UP000092553 UP000001070 UP000091820 UP000078492 UP000285301 UP000000803 UP000001292 UP000092443 UP000092460 UP000092445 UP000092444 UP000078200 UP000008711
UP000075884 UP000007819 UP000076408 UP000075883 UP000075901 UP000008237 UP000075881 UP000053097 UP000279307 UP000075920 UP000075885 UP000000673 UP000000311 UP000235965 UP000075903 UP000007062 UP000075902 UP000075840 UP000076407 UP000075882 UP000095300 UP000078540 UP000005205 UP000078541 UP000027135 UP000030765 UP000001819 UP000008744 UP000075880 UP000268350 UP000053825 UP000007755 UP000075809 UP000076502 UP000009192 UP000075900 UP000008820 UP000002320 UP000005203 UP000075886 UP000002358 UP000009046 UP000053105 UP000242457 UP000095301 UP000037069 UP000215335 UP000015103 UP000008792 UP000078542 UP000002282 UP000092553 UP000001070 UP000091820 UP000078492 UP000285301 UP000000803 UP000001292 UP000092443 UP000092460 UP000092445 UP000092444 UP000078200 UP000008711
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2H1WK07
A0A194RCB9
S4NIZ4
A0A3S2PI88
A0A194PXE6
A0A212EH75
+ More
A0A0L7LD84 A0A182FUW4 A0A182MYI3 A0A2M4AUL5 J9JQ79 A0A2M4BTZ1 A0A182Y1U6 A0A182MTN9 A0A182SCX5 E2BWX7 A0A182JNY9 A0A026WSJ5 A0A182W978 A0A182PBP9 W5JCI4 E2AKI6 A0A2J7R192 A0A182USA6 Q7Q059 A0A182TWZ7 A0A182HQ12 A0A182XD82 A0A182KSP9 A0A1I8PEQ2 A0A195BHN7 A0A1B6DT90 A0A158NJW3 A0A195F414 A0A067R026 A0A084WHY4 Q2M024 B4H8Y6 A0A182JJF8 A0A3B0J955 A0A0L7QNF3 F4WIN5 A0A151XE27 A0A154PMB8 B4KZP2 A0A182R5F8 Q172X0 B0WTM0 A0A1L8D9E0 A0A088A7H5 A0A1L8D8Z9 A0A182QDF4 K7J017 A0A1S4FG33 E0W1N2 A0A0M8ZQN4 A0A1W7R551 A0A2A3EDA6 A0A1I8MKQ1 A0A0L0BVE1 A0A232EPW0 A0A1Q3F9F6 T1PIW8 T1HYQ3 B4LCH1 A0A151I852 A0A069DYL5 A0A0P4W2X8 A0A0K8TQ39 B4PHE2 A0A1B6FGK0 A0A224XNL5 W8C2H0 A0A0M4ELX5 E9IMF5 B4J177 A0A1A9WEM5 A0A023F438 A0A336MJW5 A0A1B6KHV0 A0A0K8TX67 A0A195DV96 A0A0K8VXS7 A0A3S3PIU9 Q9VU64 A0A2S2QVY9 A0A1I8PEZ9 A0A0C9Q1K6 A0A0K8W8Q5 B4HGS0 A0A1A9Y0W9 A0A1B0B6V8 A0A1A9Y0W8 A0A1B0AI87 Q8IQJ7 D3TNA2 A0A1A9V3T8 B3NHE3 Q6AWL9 L7M380
A0A0L7LD84 A0A182FUW4 A0A182MYI3 A0A2M4AUL5 J9JQ79 A0A2M4BTZ1 A0A182Y1U6 A0A182MTN9 A0A182SCX5 E2BWX7 A0A182JNY9 A0A026WSJ5 A0A182W978 A0A182PBP9 W5JCI4 E2AKI6 A0A2J7R192 A0A182USA6 Q7Q059 A0A182TWZ7 A0A182HQ12 A0A182XD82 A0A182KSP9 A0A1I8PEQ2 A0A195BHN7 A0A1B6DT90 A0A158NJW3 A0A195F414 A0A067R026 A0A084WHY4 Q2M024 B4H8Y6 A0A182JJF8 A0A3B0J955 A0A0L7QNF3 F4WIN5 A0A151XE27 A0A154PMB8 B4KZP2 A0A182R5F8 Q172X0 B0WTM0 A0A1L8D9E0 A0A088A7H5 A0A1L8D8Z9 A0A182QDF4 K7J017 A0A1S4FG33 E0W1N2 A0A0M8ZQN4 A0A1W7R551 A0A2A3EDA6 A0A1I8MKQ1 A0A0L0BVE1 A0A232EPW0 A0A1Q3F9F6 T1PIW8 T1HYQ3 B4LCH1 A0A151I852 A0A069DYL5 A0A0P4W2X8 A0A0K8TQ39 B4PHE2 A0A1B6FGK0 A0A224XNL5 W8C2H0 A0A0M4ELX5 E9IMF5 B4J177 A0A1A9WEM5 A0A023F438 A0A336MJW5 A0A1B6KHV0 A0A0K8TX67 A0A195DV96 A0A0K8VXS7 A0A3S3PIU9 Q9VU64 A0A2S2QVY9 A0A1I8PEZ9 A0A0C9Q1K6 A0A0K8W8Q5 B4HGS0 A0A1A9Y0W9 A0A1B0B6V8 A0A1A9Y0W8 A0A1B0AI87 Q8IQJ7 D3TNA2 A0A1A9V3T8 B3NHE3 Q6AWL9 L7M380
Ontologies
GO
PANTHER
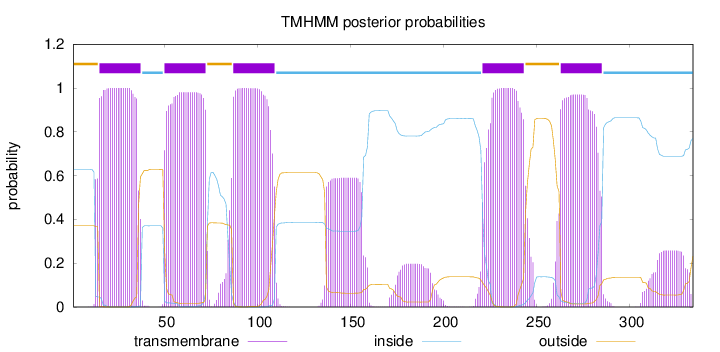
Topology
Length:
334
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
135.93775
Exp number, first 60 AAs:
33.15554
Total prob of N-in:
0.62919
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 37
inside
38 - 49
TMhelix
50 - 72
outside
73 - 86
TMhelix
87 - 109
inside
110 - 220
TMhelix
221 - 243
outside
244 - 262
TMhelix
263 - 285
inside
286 - 334
Population Genetic Test Statistics
Pi
208.505403
Theta
188.123627
Tajima's D
0.324943
CLR
0
CSRT
0.462176891155442
Interpretation
Possibly Positive selection
