Gene
KWMTBOMO04430
Pre Gene Modal
BGIBMGA005389
Annotation
PREDICTED:_putative_inorganic_phosphate_cotransporter_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.873
Sequence
CDS
ATGTCATCATTTGAAGAGAGTCCTATAATCAATGCTGATATTGAGGACGACGTTACGAATAGTCGTAGAAATCTAGTAGAAAATGAATTGGTGGAAGAAACAACTGGGTGGGTGAAATGTCGGACAGTTCTGGGTATAATGGGTTTTCTTGGTTTTGCCATAGTATATGCTATGAGAGTGAATTTATCAGTTGCAATTGTTGCCATGGTGAATTCCACTACACCCCAACCATCAAATGATTCCTCATTAGATGTATGTGAAGCCAATCCTGTTAGTAATACAACTAAACCTACAAATGGTGAATTTAATTGGACAGAACAGCAGCGCAGTCTGGTTTTAGGTTCATTCTTTTATGGTTACGTTCTAACACAGATACCGGGTGGTCGAATAGCAGAATTATATGGTGGGAAACTTGTCTATGGTTTAGGTATATTCTTCACTGCAGTATTTACAATATTAAGTCCTATTGCTGCATATACGAACTTCTGGTTCTTTATAATTGTACGAGCCCTGGCAGGATTGGCTGAAGGAGTCACATATCCTGCCATGCATGCTATGCTTGCTAAATGGATCCCACCTCTGGAAAGATCCAAGTTTGCAGCTTATGTTTATGCAGGTTCAAATATTGGAACAGTGGTCACATTACCAATATCAGGGTGGCTATGCACACTAGAGTTTGCAGGTGGATGGCCACTCTGTTTTTACATTTTCGGTGGACTCGGAGTGGTGTGGTTTATTGGATGGCTCTTCCTCATCTACGATTCACCACAAAAACACCCTAGGATATGTCCTAAAGAAATTGAATATATTGCAGGAAGCATAGGAAATGCAGAGGAAGAAAAACCCTCAATGCCGATACCATGGTGCAAGTTCCTGACGTGCGTTCCGCTGTGGGCCATCCTGATCGCGCAATGCGGACAATCGTGGGTCTTCTACACTCAGTTTACAGAAATGCCTACTTATATGGACAATATTCTGCATTTCGATATAGTATCTAACGCTGAACTGAGTGCGCTGCCGTATTTAACGGCTTGGATCGGCGGTGTGGTTCTGTGCACGATCGCGGACTGGACTATTGCTAAAGGATGGATCAGCCGACTCAATAGTATGAAACTCTGGAATACTGTTGCTGCGTTCTTGCCTGCGTTCGGCATCCTCGGCATCGCGTGGGCCGGCTGCGACAAGTTTGCGGTCATGTTGCTGCTAAGTGCAGCCTCGGCTTTCGGCGGAGCGTCTTATGCGGGTAACCAAATGAACCATATAGATCTGTCGCCGCAGTTCGCGGGCACGATGTACGGTATAACGAACGCAGCTAGCAACGTGTGCGGCTTCATGGCGCCGTACGTTGTCGGGCACATTATAAAGACAGATCAGCAAACCCTGGGTCAGTGGCGTGAAGTCTTTTATCTCGCGGCCGCTATCGATCTCGGAGCGAACTTGTTCTATTTGTTCTTCGCCAGTACCGAGGAGCAGGATTGGTCACGTCCGGACAACGACGACATGACAGCATCAGCCCCGGTCGAAAGTATTTTGTTCCCAGAGTCGTAG
Protein
MSSFEESPIINADIEDDVTNSRRNLVENELVEETTGWVKCRTVLGIMGFLGFAIVYAMRVNLSVAIVAMVNSTTPQPSNDSSLDVCEANPVSNTTKPTNGEFNWTEQQRSLVLGSFFYGYVLTQIPGGRIAELYGGKLVYGLGIFFTAVFTILSPIAAYTNFWFFIIVRALAGLAEGVTYPAMHAMLAKWIPPLERSKFAAYVYAGSNIGTVVTLPISGWLCTLEFAGGWPLCFYIFGGLGVVWFIGWLFLIYDSPQKHPRICPKEIEYIAGSIGNAEEEKPSMPIPWCKFLTCVPLWAILIAQCGQSWVFYTQFTEMPTYMDNILHFDIVSNAELSALPYLTAWIGGVVLCTIADWTIAKGWISRLNSMKLWNTVAAFLPAFGILGIAWAGCDKFAVMLLLSAASAFGGASYAGNQMNHIDLSPQFAGTMYGITNAASNVCGFMAPYVVGHIIKTDQQTLGQWREVFYLAAAIDLGANLFYLFFASTEEQDWSRPDNDDMTASAPVESILFPES
Summary
Uniprot
H9J797
A0A2H1WJX3
A0A194PWJ5
A0A194RD96
A0A212EH83
A0A0L7LD84
+ More
D6WXY7 N6UD52 A0A1Y1M0D3 U4UD81 A0A1A9WA66 A0A2J7QRJ4 A0A1A9Z8K3 W8BAI4 A0A067QQC6 A0A1A9XY00 A0A1B0BY04 A0A0K8VGU1 A0A336M9M9 A0A1L8DEP8 A0A232ETR0 A0A1L8DEJ6 K7IQ69 U5EU99 A0A1L8DEV8 A0A0A1WNA6 B4R421 T1PA44 A0A1I8MJ08 A0A1W4V7D3 B4IGM9 B3NWS3 Q9VYG7 A0A034WMJ3 A0A0M4EJ62 B4Q214 A0A1L8DEA9 A0A1L8DE08 A0A1L8DE14 Q29H64 A0A1B0GLR8 A0A0C9RKY9 B4M836 E1ZYY8 A0A158NFW9 A0A026W9G2 A0A195B2N2 F4X6V5 V9I9D3 A0A087ZVT7 E9J662 A0A195E7S4 A0A3B0JCG2 E2BP18 A0A0J7KY27 B3MRN5 A0A2A3EL64 B4L6G2 A0A151XC96 A0A195F264 A0A1J1I0M6 A0A310SM97 A0A195CHD1 A0A0L7QK51 A0A0T6BAP5 A0A154NYG1 A0A1B0FI92 Q7YTZ7 A0A0P5PS71 A0A0P6F613 A0A0P6AR80 A0A0P5EII8 A0A0P4ZUG3 A0A0P5DAU7 A0A0P5DK07 A0A0P5KC73 A0A0P5VN73 A0A0P6C5N4 A0A0P5NQ04 A0A0P5TUW6 A0A0P5A923 A0A0P5P7I4 A0A0P6JCZ1 A0A0P5Q8G0 A0A0P5ZC17 A0A0P4XIP8 A0A0P5MDG9 A0A0P5Z7V2 A0A0N7ZUR3 A0A0N8AEK6 A0A0P4ZQ45 A0A0P4WJ10 A0A0P4WRJ7 A0A0P5HE71 A0A0P5C792 A0A0P5K2Y0 A0A0P5DBP5 A0A0P5MJF9
D6WXY7 N6UD52 A0A1Y1M0D3 U4UD81 A0A1A9WA66 A0A2J7QRJ4 A0A1A9Z8K3 W8BAI4 A0A067QQC6 A0A1A9XY00 A0A1B0BY04 A0A0K8VGU1 A0A336M9M9 A0A1L8DEP8 A0A232ETR0 A0A1L8DEJ6 K7IQ69 U5EU99 A0A1L8DEV8 A0A0A1WNA6 B4R421 T1PA44 A0A1I8MJ08 A0A1W4V7D3 B4IGM9 B3NWS3 Q9VYG7 A0A034WMJ3 A0A0M4EJ62 B4Q214 A0A1L8DEA9 A0A1L8DE08 A0A1L8DE14 Q29H64 A0A1B0GLR8 A0A0C9RKY9 B4M836 E1ZYY8 A0A158NFW9 A0A026W9G2 A0A195B2N2 F4X6V5 V9I9D3 A0A087ZVT7 E9J662 A0A195E7S4 A0A3B0JCG2 E2BP18 A0A0J7KY27 B3MRN5 A0A2A3EL64 B4L6G2 A0A151XC96 A0A195F264 A0A1J1I0M6 A0A310SM97 A0A195CHD1 A0A0L7QK51 A0A0T6BAP5 A0A154NYG1 A0A1B0FI92 Q7YTZ7 A0A0P5PS71 A0A0P6F613 A0A0P6AR80 A0A0P5EII8 A0A0P4ZUG3 A0A0P5DAU7 A0A0P5DK07 A0A0P5KC73 A0A0P5VN73 A0A0P6C5N4 A0A0P5NQ04 A0A0P5TUW6 A0A0P5A923 A0A0P5P7I4 A0A0P6JCZ1 A0A0P5Q8G0 A0A0P5ZC17 A0A0P4XIP8 A0A0P5MDG9 A0A0P5Z7V2 A0A0N7ZUR3 A0A0N8AEK6 A0A0P4ZQ45 A0A0P4WJ10 A0A0P4WRJ7 A0A0P5HE71 A0A0P5C792 A0A0P5K2Y0 A0A0P5DBP5 A0A0P5MJF9
Pubmed
19121390
26354079
22118469
26227816
18362917
19820115
+ More
23537049 28004739 24495485 24845553 28648823 20075255 25830018 17994087 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 17550304 15632085 20798317 21347285 24508170 30249741 21719571 21282665 18057021
23537049 28004739 24495485 24845553 28648823 20075255 25830018 17994087 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 17550304 15632085 20798317 21347285 24508170 30249741 21719571 21282665 18057021
EMBL
BABH01013620
ODYU01009153
SOQ53380.1
KQ459589
KPI97692.1
KQ460397
+ More
KPJ15250.1 AGBW02014942 OWR40838.1 JTDY01001577 KOB73478.1 KQ971362 EFA08919.1 APGK01040126 KB740975 ENN76567.1 GEZM01043148 GEZM01043147 JAV79273.1 KB632267 ERL91012.1 NEVH01011894 PNF31193.1 GAMC01010863 JAB95692.1 KK853581 KDR06530.1 JXJN01022449 GDHF01014203 JAI38111.1 UFQT01000753 SSX26986.1 GFDF01009219 JAV04865.1 NNAY01002243 OXU21743.1 GFDF01009218 JAV04866.1 GANO01002417 JAB57454.1 GFDF01009220 JAV04864.1 GBXI01014297 JAC99994.1 CM000366 EDX17788.1 KA645607 AFP60236.1 CH480836 EDW48979.1 CH954180 EDV47235.1 AE014298 BT133363 AAF48230.1 AFH36348.1 AHN59658.1 AHN59659.1 GAKP01003949 JAC55003.1 CP012528 ALC49104.1 CM000162 EDX01535.1 GFDF01009394 JAV04690.1 GFDF01009393 JAV04691.1 GFDF01009392 JAV04692.1 CH379064 EAL31894.2 AJWK01014691 GBYB01008860 JAG78627.1 CH940653 EDW62312.1 GL435242 EFN73678.1 ADTU01014820 KK107373 QOIP01000004 EZA51649.1 RLU23294.1 KQ976662 KYM78540.1 GL888818 EGI57887.1 JR037329 AEY57718.1 GL768221 EFZ11760.1 KQ979568 KYN20874.1 OUUW01000003 SPP77772.1 GL449511 EFN82610.1 LBMM01002177 KMQ95179.1 CH902622 EDV34440.1 KPU75141.1 KZ288219 PBC32234.1 CH933812 EDW05958.2 KRG07103.1 KQ982314 KYQ57918.1 KQ981864 KYN34262.1 CVRI01000026 CRK92382.1 KQ763450 OAD55056.1 KQ977754 KYN00113.1 KQ415041 KOC58896.1 LJIG01002547 KRT84393.1 KQ434782 KZC04621.1 CCAG010018157 CCAG010018158 BT010092 AAQ22561.1 GDIQ01124326 JAL27400.1 GDIQ01251248 GDIQ01249698 GDIQ01227001 GDIQ01203300 GDIQ01189313 GDIQ01139338 GDIQ01070551 GDIQ01067513 GDIQ01052814 JAN27224.1 GDIP01025660 JAM78055.1 GDIP01159548 JAJ63854.1 GDIP01209487 GDIP01134843 LRGB01003123 JAJ13915.1 KZS04440.1 GDIP01160390 JAJ63012.1 GDIP01161189 JAJ62213.1 GDIQ01212149 JAK39576.1 GDIP01097758 JAM05957.1 GDIP01020617 GDIQ01081904 JAM83098.1 JAN12833.1 GDIQ01139337 GDIQ01136879 JAL12389.1 GDIP01124148 GDIQ01081906 GDIQ01078698 JAL79566.1 JAN12831.1 GDIP01203389 JAJ20013.1 GDIQ01136882 JAL14844.1 GDIQ01009078 JAN85659.1 GDIQ01124327 JAL27399.1 GDIP01046314 JAM57401.1 GDIP01240647 JAI82754.1 GDIQ01161312 JAK90413.1 GDIP01047850 JAM55865.1 GDIP01210633 JAJ12769.1 GDIP01155089 JAJ68313.1 GDIP01209488 JAJ13914.1 GDRN01042058 JAI67078.1 GDRN01042061 GDRN01042060 GDRN01042059 JAI67077.1 GDIQ01234946 JAK16779.1 GDIP01174590 JAJ48812.1 GDIQ01195407 JAK56318.1 GDIP01174588 JAJ48814.1 GDIQ01155713 JAK96012.1
KPJ15250.1 AGBW02014942 OWR40838.1 JTDY01001577 KOB73478.1 KQ971362 EFA08919.1 APGK01040126 KB740975 ENN76567.1 GEZM01043148 GEZM01043147 JAV79273.1 KB632267 ERL91012.1 NEVH01011894 PNF31193.1 GAMC01010863 JAB95692.1 KK853581 KDR06530.1 JXJN01022449 GDHF01014203 JAI38111.1 UFQT01000753 SSX26986.1 GFDF01009219 JAV04865.1 NNAY01002243 OXU21743.1 GFDF01009218 JAV04866.1 GANO01002417 JAB57454.1 GFDF01009220 JAV04864.1 GBXI01014297 JAC99994.1 CM000366 EDX17788.1 KA645607 AFP60236.1 CH480836 EDW48979.1 CH954180 EDV47235.1 AE014298 BT133363 AAF48230.1 AFH36348.1 AHN59658.1 AHN59659.1 GAKP01003949 JAC55003.1 CP012528 ALC49104.1 CM000162 EDX01535.1 GFDF01009394 JAV04690.1 GFDF01009393 JAV04691.1 GFDF01009392 JAV04692.1 CH379064 EAL31894.2 AJWK01014691 GBYB01008860 JAG78627.1 CH940653 EDW62312.1 GL435242 EFN73678.1 ADTU01014820 KK107373 QOIP01000004 EZA51649.1 RLU23294.1 KQ976662 KYM78540.1 GL888818 EGI57887.1 JR037329 AEY57718.1 GL768221 EFZ11760.1 KQ979568 KYN20874.1 OUUW01000003 SPP77772.1 GL449511 EFN82610.1 LBMM01002177 KMQ95179.1 CH902622 EDV34440.1 KPU75141.1 KZ288219 PBC32234.1 CH933812 EDW05958.2 KRG07103.1 KQ982314 KYQ57918.1 KQ981864 KYN34262.1 CVRI01000026 CRK92382.1 KQ763450 OAD55056.1 KQ977754 KYN00113.1 KQ415041 KOC58896.1 LJIG01002547 KRT84393.1 KQ434782 KZC04621.1 CCAG010018157 CCAG010018158 BT010092 AAQ22561.1 GDIQ01124326 JAL27400.1 GDIQ01251248 GDIQ01249698 GDIQ01227001 GDIQ01203300 GDIQ01189313 GDIQ01139338 GDIQ01070551 GDIQ01067513 GDIQ01052814 JAN27224.1 GDIP01025660 JAM78055.1 GDIP01159548 JAJ63854.1 GDIP01209487 GDIP01134843 LRGB01003123 JAJ13915.1 KZS04440.1 GDIP01160390 JAJ63012.1 GDIP01161189 JAJ62213.1 GDIQ01212149 JAK39576.1 GDIP01097758 JAM05957.1 GDIP01020617 GDIQ01081904 JAM83098.1 JAN12833.1 GDIQ01139337 GDIQ01136879 JAL12389.1 GDIP01124148 GDIQ01081906 GDIQ01078698 JAL79566.1 JAN12831.1 GDIP01203389 JAJ20013.1 GDIQ01136882 JAL14844.1 GDIQ01009078 JAN85659.1 GDIQ01124327 JAL27399.1 GDIP01046314 JAM57401.1 GDIP01240647 JAI82754.1 GDIQ01161312 JAK90413.1 GDIP01047850 JAM55865.1 GDIP01210633 JAJ12769.1 GDIP01155089 JAJ68313.1 GDIP01209488 JAJ13914.1 GDRN01042058 JAI67078.1 GDRN01042061 GDRN01042060 GDRN01042059 JAI67077.1 GDIQ01234946 JAK16779.1 GDIP01174590 JAJ48812.1 GDIQ01195407 JAK56318.1 GDIP01174588 JAJ48814.1 GDIQ01155713 JAK96012.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000037510
UP000007266
+ More
UP000019118 UP000030742 UP000091820 UP000235965 UP000092445 UP000027135 UP000092443 UP000092460 UP000215335 UP000002358 UP000000304 UP000095301 UP000192221 UP000001292 UP000008711 UP000000803 UP000092553 UP000002282 UP000001819 UP000092461 UP000008792 UP000000311 UP000005205 UP000053097 UP000279307 UP000078540 UP000007755 UP000005203 UP000078492 UP000268350 UP000008237 UP000036403 UP000007801 UP000242457 UP000009192 UP000075809 UP000078541 UP000183832 UP000078542 UP000053825 UP000076502 UP000092444 UP000076858
UP000019118 UP000030742 UP000091820 UP000235965 UP000092445 UP000027135 UP000092443 UP000092460 UP000215335 UP000002358 UP000000304 UP000095301 UP000192221 UP000001292 UP000008711 UP000000803 UP000092553 UP000002282 UP000001819 UP000092461 UP000008792 UP000000311 UP000005205 UP000053097 UP000279307 UP000078540 UP000007755 UP000005203 UP000078492 UP000268350 UP000008237 UP000036403 UP000007801 UP000242457 UP000009192 UP000075809 UP000078541 UP000183832 UP000078542 UP000053825 UP000076502 UP000092444 UP000076858
Interpro
CDD
ProteinModelPortal
H9J797
A0A2H1WJX3
A0A194PWJ5
A0A194RD96
A0A212EH83
A0A0L7LD84
+ More
D6WXY7 N6UD52 A0A1Y1M0D3 U4UD81 A0A1A9WA66 A0A2J7QRJ4 A0A1A9Z8K3 W8BAI4 A0A067QQC6 A0A1A9XY00 A0A1B0BY04 A0A0K8VGU1 A0A336M9M9 A0A1L8DEP8 A0A232ETR0 A0A1L8DEJ6 K7IQ69 U5EU99 A0A1L8DEV8 A0A0A1WNA6 B4R421 T1PA44 A0A1I8MJ08 A0A1W4V7D3 B4IGM9 B3NWS3 Q9VYG7 A0A034WMJ3 A0A0M4EJ62 B4Q214 A0A1L8DEA9 A0A1L8DE08 A0A1L8DE14 Q29H64 A0A1B0GLR8 A0A0C9RKY9 B4M836 E1ZYY8 A0A158NFW9 A0A026W9G2 A0A195B2N2 F4X6V5 V9I9D3 A0A087ZVT7 E9J662 A0A195E7S4 A0A3B0JCG2 E2BP18 A0A0J7KY27 B3MRN5 A0A2A3EL64 B4L6G2 A0A151XC96 A0A195F264 A0A1J1I0M6 A0A310SM97 A0A195CHD1 A0A0L7QK51 A0A0T6BAP5 A0A154NYG1 A0A1B0FI92 Q7YTZ7 A0A0P5PS71 A0A0P6F613 A0A0P6AR80 A0A0P5EII8 A0A0P4ZUG3 A0A0P5DAU7 A0A0P5DK07 A0A0P5KC73 A0A0P5VN73 A0A0P6C5N4 A0A0P5NQ04 A0A0P5TUW6 A0A0P5A923 A0A0P5P7I4 A0A0P6JCZ1 A0A0P5Q8G0 A0A0P5ZC17 A0A0P4XIP8 A0A0P5MDG9 A0A0P5Z7V2 A0A0N7ZUR3 A0A0N8AEK6 A0A0P4ZQ45 A0A0P4WJ10 A0A0P4WRJ7 A0A0P5HE71 A0A0P5C792 A0A0P5K2Y0 A0A0P5DBP5 A0A0P5MJF9
D6WXY7 N6UD52 A0A1Y1M0D3 U4UD81 A0A1A9WA66 A0A2J7QRJ4 A0A1A9Z8K3 W8BAI4 A0A067QQC6 A0A1A9XY00 A0A1B0BY04 A0A0K8VGU1 A0A336M9M9 A0A1L8DEP8 A0A232ETR0 A0A1L8DEJ6 K7IQ69 U5EU99 A0A1L8DEV8 A0A0A1WNA6 B4R421 T1PA44 A0A1I8MJ08 A0A1W4V7D3 B4IGM9 B3NWS3 Q9VYG7 A0A034WMJ3 A0A0M4EJ62 B4Q214 A0A1L8DEA9 A0A1L8DE08 A0A1L8DE14 Q29H64 A0A1B0GLR8 A0A0C9RKY9 B4M836 E1ZYY8 A0A158NFW9 A0A026W9G2 A0A195B2N2 F4X6V5 V9I9D3 A0A087ZVT7 E9J662 A0A195E7S4 A0A3B0JCG2 E2BP18 A0A0J7KY27 B3MRN5 A0A2A3EL64 B4L6G2 A0A151XC96 A0A195F264 A0A1J1I0M6 A0A310SM97 A0A195CHD1 A0A0L7QK51 A0A0T6BAP5 A0A154NYG1 A0A1B0FI92 Q7YTZ7 A0A0P5PS71 A0A0P6F613 A0A0P6AR80 A0A0P5EII8 A0A0P4ZUG3 A0A0P5DAU7 A0A0P5DK07 A0A0P5KC73 A0A0P5VN73 A0A0P6C5N4 A0A0P5NQ04 A0A0P5TUW6 A0A0P5A923 A0A0P5P7I4 A0A0P6JCZ1 A0A0P5Q8G0 A0A0P5ZC17 A0A0P4XIP8 A0A0P5MDG9 A0A0P5Z7V2 A0A0N7ZUR3 A0A0N8AEK6 A0A0P4ZQ45 A0A0P4WJ10 A0A0P4WRJ7 A0A0P5HE71 A0A0P5C792 A0A0P5K2Y0 A0A0P5DBP5 A0A0P5MJF9
PDB
6E9N
E-value=4.17357e-18,
Score=225
Ontologies
GO
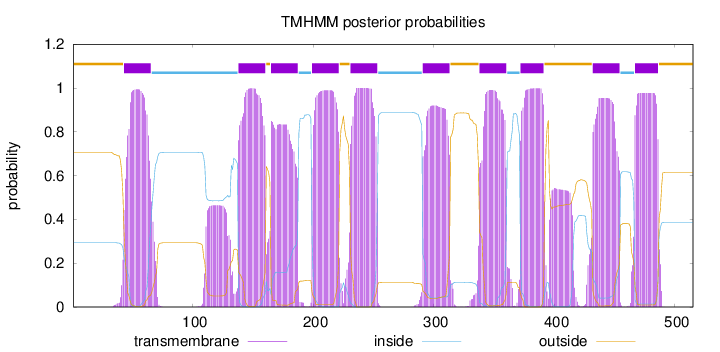
Topology
Length:
515
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
232.4559
Exp number, first 60 AAs:
16.48742
Total prob of N-in:
0.29466
POSSIBLE N-term signal
sequence
outside
1 - 42
TMhelix
43 - 65
inside
66 - 137
TMhelix
138 - 160
outside
161 - 164
TMhelix
165 - 187
inside
188 - 198
TMhelix
199 - 221
outside
222 - 230
TMhelix
231 - 253
inside
254 - 290
TMhelix
291 - 313
outside
314 - 337
TMhelix
338 - 360
inside
361 - 371
TMhelix
372 - 391
outside
392 - 431
TMhelix
432 - 454
inside
455 - 466
TMhelix
467 - 486
outside
487 - 515
Population Genetic Test Statistics
Pi
208.942463
Theta
177.052828
Tajima's D
0.580509
CLR
2.2748
CSRT
0.540022998850057
Interpretation
Possibly Positive selection
